3C5H
 
 | | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP) | | Descriptor: | Glucocorticoid receptor DNA-binding factor 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Ras homolog domain of human GRLF1 (p190RhoGAP).
To be Published
|
|
3BNP
 
 | |
4FVD
 
 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4NG4
 
 | | Structure of phosphoglycerate kinase (CBU_1782) from Coxiella burnetii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoglycerate kinase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2013-11-01 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3BM4
 
 | | Crystal Structure of Human ADP-ribose Pyrophosphatase NUDT5 In complex with magnesium and AMPcpr | | Descriptor: | ADP-sugar pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MAGNESIUM ION | | Authors: | Zha, M, Guo, Q, Zhang, Y, Zhong, C, Ou, Y, Ding, J. | | Deposit date: | 2007-12-12 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of ADP-Ribose Hydrolysis By Human NUDT5 From Structural and Kinetic Studies
J.Mol.Biol., 379, 2008
|
|
2CIV
 
 | | Chloroperoxidase bromide complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CHLOROPEROXIDASE, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | Authors: | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | Deposit date: | 2016-11-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (7.086 Å) | | Cite: | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
5U2B
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the phenylamino-substituted estrogen, (8R,9S,13S,14S,17S)-13-methyl-17-(phenylamino)-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol, without a coactivator peptide | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},17~{S})-13-methyl-17-phenylazanyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthren-3-ol, Estrogen receptor | | Authors: | Nwachukwu, J.C, Nowak, J, Carlson, K.E, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Molecular Mechanisms of Cytokine-Mediated Endocrine Resistance in Human Breast Cancer Cells.
Mol. Cell, 65, 2017
|
|
3BZ5
 
 | | Functional domain of InlJ from Listeria monocytogenes includes a cysteine ladder | | Descriptor: | CHLORIDE ION, Internalin-J, SULFATE ION | | Authors: | Bublitz, M, Holland, C, Sabet, C, Reichelt, J, Cossart, P, Heinz, D.W, Bierne, H, Schubert, W.D. | | Deposit date: | 2008-01-17 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and standardized geometric analysis of InlJ, a listerial virulence factor and leucine-rich repeat protein with a novel cysteine ladder.
J.Mol.Biol., 378, 2008
|
|
2CKC
 
 | | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | | Descriptor: | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 7 | | Authors: | Ab, E, de Jong, R.N, Diercks, T, Xiaoyun, J, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction
To be Published
|
|
3BUY
 
 | | MHC-I in complex with peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Rossjohn, J, La Gruta, N.L, Purcell, A.W, Turner, S.J, Dunstone, M.A. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5U16
 
 | | Structure of human MR1-2-OH-1-NA in complex with human MAIT A-F7 TCR | | Descriptor: | 2-hydroxynaphthalene-1-carbaldehyde, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Keller, A.N, Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2016-11-27 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells.
Nat. Immunol., 18, 2017
|
|
2CVX
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3BNT
 
 | |
5UCK
 
 | | Class II fructose-1,6-bisphosphate aldolase of Helicobacter pylori with cleavage products | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
3BQC
 
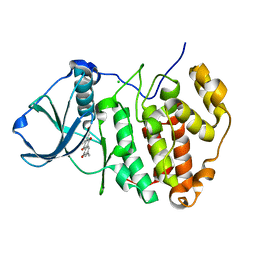 | | High pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
2GIE
 
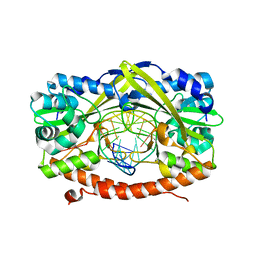 | | HincII bound to cognate DNA GTTAAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*G)-3', SODIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
3BRU
 
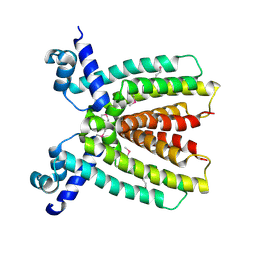 | |
3C1P
 
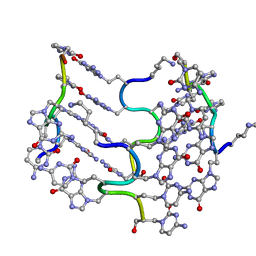 | | Crystal Structure of an alternating D-Alanyl, L-Homoalanyl PNA | | Descriptor: | Peptide Nucleic Acid DLY-HGL-AGD-LHC-AGD-LHC-CUD-LYS | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M, Zhang, J, Diederichsen, U. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Continuous beta-turn fold of an alternating alanyl/homoalanyl peptide nucleic acid.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2CN0
 
 | | Complex of Recombinant Human Thrombin with a Designed Inhibitor | | Descriptor: | 4-(1R,3AS,4R,8AS,8BR)-[1-DIFLUOROMETHYL-2-(4-FLUOROBENZYL)-3-OXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZAMIDINE, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Hoffmann-Roder, A, Schweizer, E, Egger, J, Seiler, P, Obst-Sander, U, Wagner, B, Kansy, M, Banner, D.W, Diederich, F. | | Deposit date: | 2006-05-17 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mapping the Fluorophilicity of a Hydrophobic Pocket: Synthesis and Biological Evaluation of Tricyclic Thrombin Inhibitors Directing Fluorinated Alkyl Groups Into the P Pocket
Chemmedchem, 1, 2006
|
|
3BT6
 
 | | Crystal Structure of Influenza B Virus Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q, Cheng, F, Lu, M, Tian, X, Ma, J. | | Deposit date: | 2007-12-27 | | Release date: | 2008-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of unliganded influenza B virus hemagglutinin.
J.Virol., 82, 2008
|
|
2CSL
 
 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
2CVV
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonucleoside-diphosphate reductase large chain 1, ... | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2CVT
 
 | | Structures of Yeast Ribonucleotide Reductase I | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1QU6
 
 | | STRUCTURE OF THE DOUBLE-STRANDED RNA-BINDING DOMAIN OF THE PROTEIN KINASE PKR REVEALS THE MOLECULAR BASIS OF ITS DSRNA-MEDIATED ACTIVATION | | Descriptor: | PROTEIN KINASE PKR | | Authors: | Nanduri, S, Carpick, B.W, Yang, Y, Williams, B.R.G, Qin, J. | | Deposit date: | 1999-07-08 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation.
EMBO J., 17, 1998
|
|
