8C1J
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Protein arginine N-methyltransferase 2, RNA-binding protein Rsf1-like | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_18-30
To Be Published
|
|
5XB2
 
 | | ADP-Mg-F-dTMP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.161 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate
FEBS J., 284, 2017
|
|
1ZJY
 
 | | Structure of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis in complex with phenylethanol and NADH | | Descriptor: | (1R)-1-PHENYLETHANOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
6GR7
 
 | | Crystal Structure Of Human Transthyretin in complex with 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) | | Descriptor: | 2-[2,4,5-tris(chloranyl)phenoxy]ethanoic acid, Transthyretin | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
3QQ9
 
 | | Crystal structure of FAB fragment of anti-human RSV (RESPIRATORY SYNCYTIAL VIRUS) F Protein MAB 101F | | Descriptor: | 101F HEAVY CHAIN, 101F LIGHT CHAIN, SULFATE ION | | Authors: | Luo, J, Tsui, P, Spurlino, J, Lewansowski, F, Heavner, G.A, Del Vecchio, F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Fab 101F
To be Published
|
|
7VKK
 
 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
5I0C
 
 | | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12 | | Descriptor: | CADMIUM ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of predicted acyltransferase YjdJ with acyl-CoA N-acyltransferase domain from Escherichia coli str. K-12
To Be Published
|
|
5I1L
 
 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV5-51/IGKV4-1 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, GLYCEROL | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
2JAF
 
 | | Ground state of halorhodopsin T203V | | Descriptor: | CHLORIDE ION, Halorhodopsin, PALMITIC ACID, ... | | Authors: | Gmelin, W, Zeth, K, Efremov, R, Heberle, J, Tittor, J, Oesterhelt, D. | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 intermediate of halorhodopsin at 1.9 angstroms resolution.
Photochem. Photobiol., 83, 2007
|
|
5IQ2
 
 | |
8CD1
 
 | | 70S-PHIKZ014 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Gerovac, M, Vogel, J. | | Deposit date: | 2023-01-29 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Phage proteins target and co-opt host ribosomes immediately upon infection.
Nat Microbiol, 9, 2024
|
|
7VIL
 
 | |
7VKR
 
 | |
7PD7
 
 | | Crocagin methyl transferase CgnL | | Descriptor: | GLYCEROL, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zheng, D, Koehnke, J. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
3QLE
 
 | | Structural Basis for the Function of Tim50 in the Mitochondrial Presequence Translocase | | Descriptor: | ACETATE ION, CALCIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Qian, X.G, Gebert, M, Hpker, J, Yan, M, Li, J.Z, Wiedemann, N, Laan, M.V.D, Pfanner, N, Sha, B.D. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Structural basis for the function of tim50 in the mitochondrial presequence translocase.
J.Mol.Biol., 411, 2011
|
|
3QYC
 
 | | Structure of a dimeric anti-HER2 single domain antibody | | Descriptor: | VH domain of IgG molecule | | Authors: | Baral, T.N, Chao, S, Li, S, Tanha, J, Arbabai, M, Wang, S, Zhang, J. | | Deposit date: | 2011-03-03 | | Release date: | 2012-02-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Human Single Domain Antibody Dimer Formed through V(H)-V(H) Non-Covalent Interactions.
Plos One, 7, 2012
|
|
8BY6
 
 | | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
2N52
 
 | |
7VKQ
 
 | |
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
8SMG
 
 | | Structure of SPO1 phage Tad2 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
5IDL
 
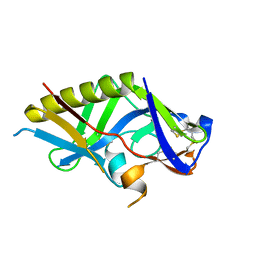 | | Crystal structure of the germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Germline-targeting HIV-1 gp120 engineered outer domain, eOD-GT8 | | Authors: | Julien, J.P, Ereno-Orbea, J, Jardine, J.G, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-02-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen.
Science, 351, 2016
|
|
5XJO
 
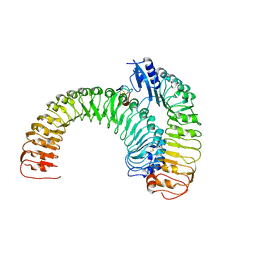 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
5I17
 
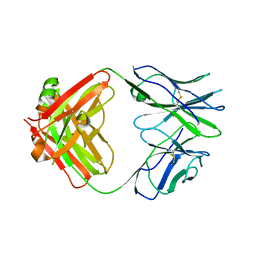 | | CRYSTAL STRUCTURE OF HUMAN GERMLINE ANTIBODY IGHV1-69/IGKV3-20 | | Descriptor: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Luo, J, Gilliland, G. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-08 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural diversity in a human antibody germline library.
Mabs, 8, 2016
|
|
3IV2
 
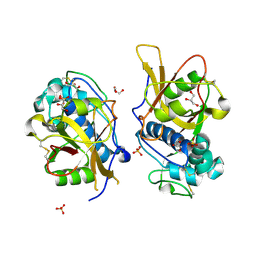 | | Crystal structure of mature apo-Cathepsin L C25A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, GLYCEROL, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J. | | Deposit date: | 2009-08-31 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
