8F8X
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
7A60
 
 | | Crystal structure of VIM-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A61
 
 | | Crystal structure of KPC-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (2~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-butyl-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A63
 
 | | Crystal structure of L1 with hydrolyzed faropenem (imine, ring-closed form) | | Descriptor: | (2R,5S)-2-[(1S,2R)-1-carboxy-2-hydroxy-propyl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydrothiazole-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57000113 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A6P
 
 | |
6L1F
 
 | |
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
6WXL
 
 | |
6L84
 
 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
8T12
 
 | |
8T13
 
 | |
4PW2
 
 | | Crystal structure of D-glucuronyl C5 epimerase | | Descriptor: | CITRIC ACID, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
6UIY
 
 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-{[(pyridin-2-yl)methyl][(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)pentanamide}iron(2+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
8CO4
 
 | | Crystal structure of apo S-nitrosoglutathione reductase from Arabidopsis thalina | | Descriptor: | 1,2-ETHANEDIOL, Alcohol dehydrogenase class-3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Fanti, S, Carloni, G, Rossi, J, Falini, G, Zaffagnini, M. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of Arabidopsis alcohol dehydrogenases reveals distinct functional properties but similar redox sensitivity.
Plant J., 118, 2024
|
|
4Q0K
 
 | | Crystal Structure of Phytohormone Binding Protein from Medicago truncatula in complex with gibberellic acid (GA3) | | Descriptor: | GIBBERELLIN A3, GLYCEROL, PHYTOHORMONE BINDING PROTEIN MTPHBP | | Authors: | Ciesielska, A, Barciszewski, J, Ruszkowski, M, Jaskolski, M, Sikorski, M. | | Deposit date: | 2014-04-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Specific binding of gibberellic acid by Cytokinin-Specific Binding Proteins: a new aspect of plant hormone-binding proteins with the PR-10 fold.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6TVQ
 
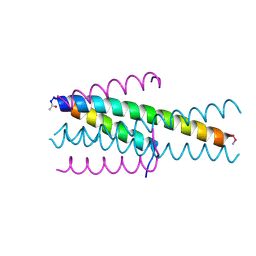 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVU
 
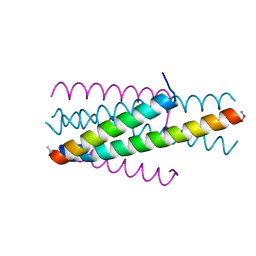 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Transmembrane protein gp41 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVW
 
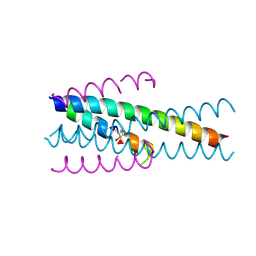 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Envelope glycoprotein, Transmembrane protein gp41,Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6LHV
 
 | | Structure of FANCA and FANCG Complex | | Descriptor: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
8GLS
 
 | |
2BXV
 
 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 2-({[3-FLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENT-1-ENE-1-CARBOXYLIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Baumgartner, R, Walloschek, M, Karlik, M, Gotschlich, A, Tasler, S, Mies, J, Leban, J. | | Deposit date: | 2005-07-27 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J. Med. Chem., 49, 2006
|
|
6ZRM
 
 | | G-quadruplex with a G-A bulge | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*AP*GP*GP*GP*AP*GP*CP*GP*GP*GP*AP*GP*TP*GP*GP*G)-3') | | Authors: | Lenarcic Zivkovic, M, Plavec, J. | | Deposit date: | 2020-07-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA G-Quadruplex Related to Osteoporosis with a G-A Bulge Forming a Pseudo-loop .
Molecules, 25, 2020
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
8CIN
 
 | | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-5 fab HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
4X09
 
 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
