3JAD
 
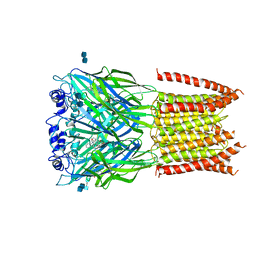 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, STRYCHNINE | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
4A27
 
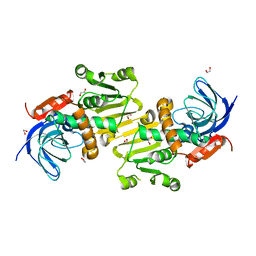 | | Crystal structure of human synaptic vesicle membrane protein VAT-1 homolog-like protein | | Descriptor: | 1,2-ETHANEDIOL, SYNAPTIC VESICLE MEMBRANE PROTEIN VAT-1 HOMOLOG-LIKE | | Authors: | Vollmar, M, Shafqat, N, Muniz, J.R.C, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Synaptic Vesicle Membrane Protein Vat-1 Homolog-Like Protein
To be Published
|
|
4AKM
 
 | |
3JVN
 
 | | Crystal Structure of the acetyltransferase VF_1542 from Vibrio fischeri, Northeast Structural Genomics Consortium Target VfR136 | | Descriptor: | Acetyltransferase, SULFATE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-09-29 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Northeast Structural Genomics Consortium Target VfR136
To be Published
|
|
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
433D
 
 | |
4KEH
 
 | | Crosslinked Crystal Structure of Type II Fatty Synthase Dehydratase, FabA, and Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, N-{3-[DIHYDROXY(NONYL)-LAMBDA~4~-SULFANYL]PROPYL}-N~3~-[(2R)-2-HYDROXY-3,3-DIMETHYL-4-(PHOSPHONOOXY)BUTANOYL]-BETA-ALANINAMIDE, N-{3-[dihydroxy(nonyl)-lambda~4~-sulfanyl]propyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Nguyen, C, Haushalter, R, Finzel, K, Leong, J, Le, B.C, Burkart, M, Tsai, S.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Trapping the dynamic acyl carrier protein in fatty acid biosynthesis.
Nature, 505, 2014
|
|
1RVH
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER GCAAAATTTTGC | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*AP*TP*TP*TP*TP*GP*C)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
458D
 
 | |
3HX6
 
 | |
3HZ1
 
 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
1R1Y
 
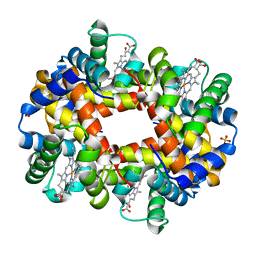 | | Crystal structure of deoxy-human hemoglobin Bassett at 1.8 angstrom | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abdulmalik, O, Safo, M.K, Lerner, N.B, Ochotorena, J, Dhaikin, Y, Abraham, D.J, Asakura, T. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of hemoglobin bassett (alpha94Asp-->Ala), a variant with very low oxygen affinity
Am.J.Hematol., 77, 2004
|
|
3HZ7
 
 | | Crystal Structure of the SirA-like protein (DSY4693) from Desulfitobacterium hafniense, Northeast Structural Genomics Consortium Target DhR2A | | Descriptor: | SULFUR OXIDE, Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target DhR2A
To be Published
|
|
1R52
 
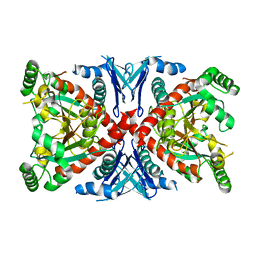 | | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae | | Descriptor: | Chorismate synthase | | Authors: | Quevillon-Cheruel, S, Leulliot, N, Meyer, P, Graille, M, Bremang, M, Blondeau, K, Sorel, I, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-10-09 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the bifunctional chorismate synthase from Saccharomyces cerevisiae
J.Biol.Chem., 279, 2004
|
|
4ABN
 
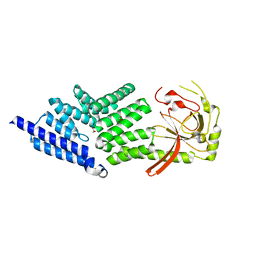 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ADD
 
 | |
3I23
 
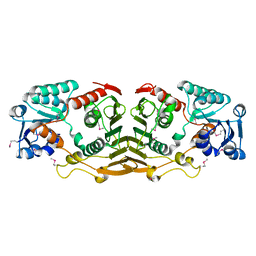 | | Crystal Structure of an Oxidoreductase (Gfo/Idh/MocA family) from Enterococcus faecalis. Northeast Structural Genomics Consortium target id EfR167 | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Oxidoreductase (Gfo/Idh/MocA family) from Enterococcus faecalis. Northeast Structural Genomics Consortium target id EfR167
To be Published
|
|
1R8H
 
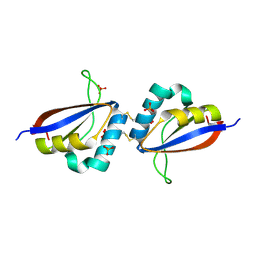 | | Comparison of the structure and DNA binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus | | Descriptor: | PHOSPHATE ION, Regulatory protein E2 | | Authors: | Dell, G, Wilkinson, K.W, Tranter, R, Parish, J, Brady, R.L, Gaston, K. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus.
J.Mol.Biol., 334, 2003
|
|
3I2R
 
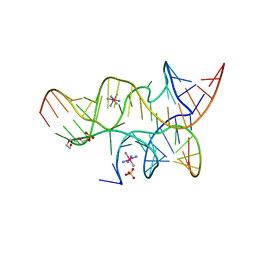 | | Crystal structure of the hairpin ribozyme with a 2',5'-linked substrate with N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
4AF8
 
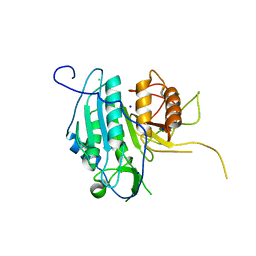 | | The structural basis for metacaspase substrate specificity and activation | | Descriptor: | CHLORIDE ION, METACASPASE MCA2, SODIUM ION | | Authors: | McLuskey, K, Rudolf, J, Isaacs, N.W, Coombs, G.H, Moss, C.X, Mottram, J.C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Trypanosoma Brucei Metacaspase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3HYY
 
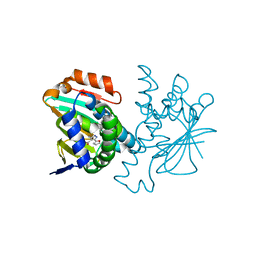 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
463D
 
 | | CALCIUM FORM OF D(CGCGAATTCGCG)2 | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Liu, J, Subirana, J.A. | | Deposit date: | 1999-03-29 | | Release date: | 1999-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of d(CGCGAATTCGCG) in the presence of Ca(2+) ions.
J.Biol.Chem., 274, 1999
|
|
4ACJ
 
 | | Crystal structure of the TLDC domain of Oxidation resistance protein 2 from zebrafish | | Descriptor: | WU:FB25H12 PROTEIN, | | Authors: | Blaise, M, B Alsarraf, H.M.A, Wong, J.E.M.M, Midtgaard, S.R, Laroche, F, Schack, L, Spaink, H, Stougaard, J, Thirup, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal Structure of the Tldc Domain of Oxidation Resistance Protein 2 from Zebrafish.
Proteins, 80, 2012
|
|
3I1I
 
 | | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis | | Descriptor: | ACETATE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis.
To be published
|
|
4MW9
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1478) | | Descriptor: | 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
