3RVB
 
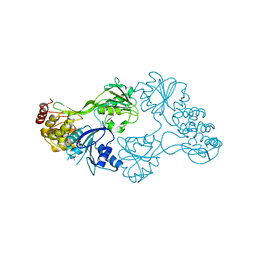 | | The structure of HCV NS3 helicase (Heli-80) bound with inhibitor ITMN-3479 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA helicase, ... | | Authors: | Mather, O, Cheng, R, Schonfield, D, Barker, J. | | Deposit date: | 2011-05-06 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of HCV NS3 helicase with inhibitor ITMN-3479
To be Published
|
|
1U31
 
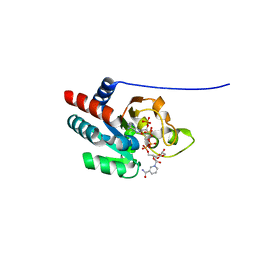 | | recombinant human heart transhydrogenase dIII bound with NADPH | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase, mitochondrial, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U2D
 
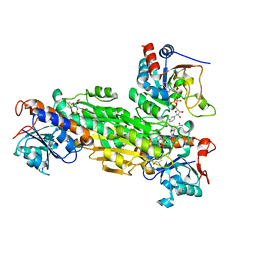 | | Structre of transhydrogenaes (dI.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U2G
 
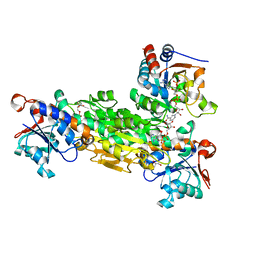 | | transhydrogenase (dI.ADPr)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U28
 
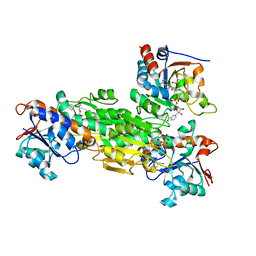 | | R. rubrum transhydrogenase asymmetric complex (dI.NAD+)2(dIII.NADP+)1 | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
2FR8
 
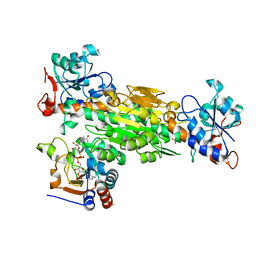 | | Structure of transhydrogenase (dI.R127A.NAD+)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
2FRD
 
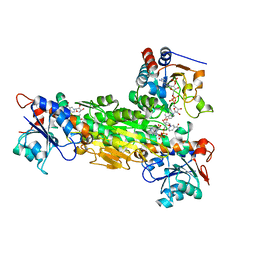 | | Structure of Transhydrogenase (dI.S138A.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
2FSV
 
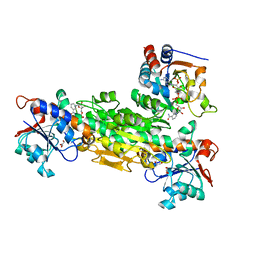 | | Structure of transhydrogenase (dI.D135N.NAD+)2(dIII.E155W.NADP+)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
3FT5
 
 | | Structure of HSP90 bound with a novel fragment | | Descriptor: | 4-methyl-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Barker, J.B, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B. | | Deposit date: | 2009-01-12 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Identification of Hsp90 Inhibitors.
Chemmedchem, 4, 2009
|
|
3HZ5
 
 | | Crystal structure of Hsp90 with fragment Z064 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(5-furan-2-yl-3-methyl-1H-pyrazol-4-yl)butyl]-N-methyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment Z064
to be published
|
|
3HYZ
 
 | | Crystal structure of Hsp90 with fragment 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragment 42-C03
TO BE PUBLISHED
|
|
3HZ1
 
 | | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03 | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Hsp90 with fragments 37-D04 and 42-C03
TO BE PUBLISHED
|
|
3HYY
 
 | | Crystal structure of Hsp90 with fragment 37-D04 | | Descriptor: | Heat shock protein HSP 90-alpha, methyl 5-furan-2-yl-3-methyl-1H-pyrazole-4-carboxylate | | Authors: | Barker, J, Mather, O, Cheng, R.K.Y, Palan, S, Felicetti, B, Whittaker, M. | | Deposit date: | 2009-06-23 | | Release date: | 2010-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Hsp90 with fragment 37-D04
TO BE PUBLISHED
|
|
3L6C
 
 | | X-ray crystal structure of rat serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Wood, M, Schonfeld, D, Mather, O, Cesura, A, Barker, J. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
