8FZF
 
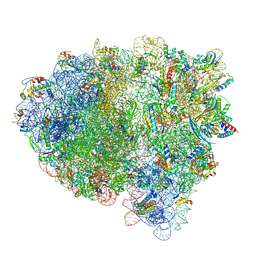 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP
Nat.Struct.Mol.Biol., 2024
|
|
8FZ9
 
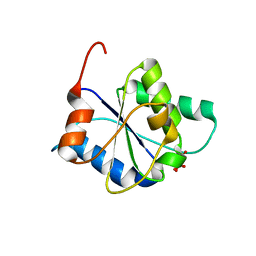 | |
4HZ2
 
 | | Crystal structure of glutathione s-transferase xaut_3756 (target efi-507152) from xanthobacter autotrophicus py2 | | Descriptor: | BENZOIC ACID, GLUTATHIONE, Glutathione S-transferase domain, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glutathione s-transferase xaut_3756 from xanthobacter autotrophicus py2
To be Published
|
|
8G2Q
 
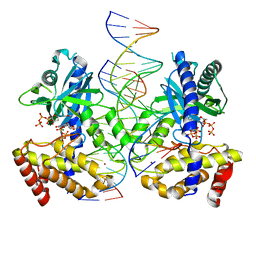 | |
8G2P
 
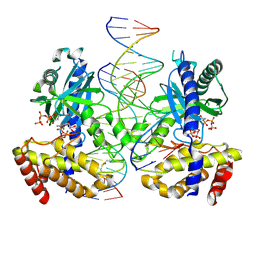 | |
8FJ4
 
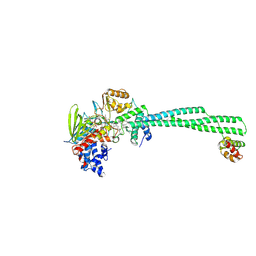 | | LSD1-CoREST in complex with T108, short soaking | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-phenylbenzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8FSE
 
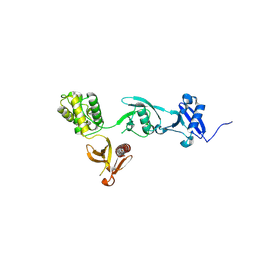 | |
8FTB
 
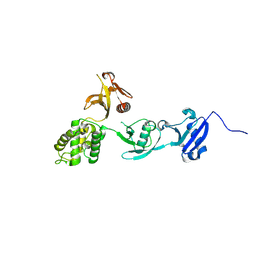 | |
8FJ6
 
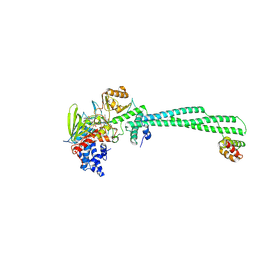 | | LSD1-CoREST in complex with T108, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[7,8-dimethyl-2,4-dioxo-5-{3-[3-(phenylcarbamoyl)phenyl]propanoyl}-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2022-12-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
7T4Q
 
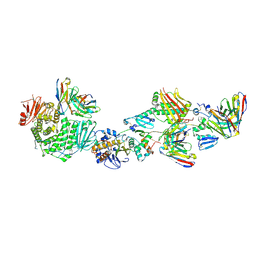 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
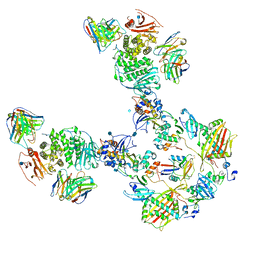 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
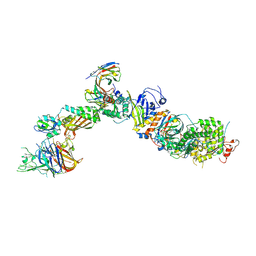 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
8GAP
 
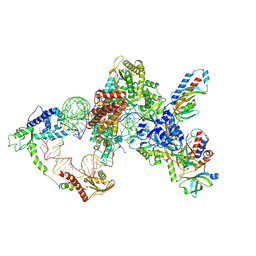 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
4EP1
 
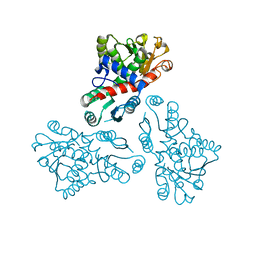 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Mikolajczak, K, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of anabolic ornithine carbamoyltransferase from Bacillus anthracis
To be Published
|
|
3EQP
 
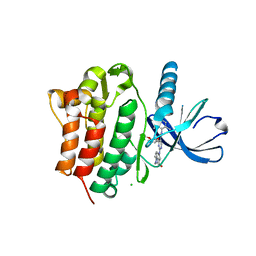 | | Crystal Structure of Ack1 with compound T95 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N-(2,6-dimethylphenyl)-4-(2-ethoxyphenoxy)-2-({4-[4-(2-hydroxyethyl)piperazin-1-yl]phenyl}amino)pyrimidine-5-carboxamide | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8GX9
 
 | |
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
3T9K
 
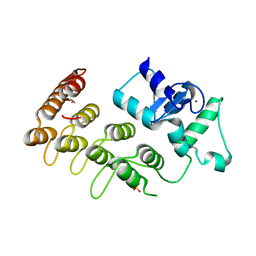 | | Crystal Structure of ACAP1 C-portion mutant S554D fused with integrin beta1 peptide | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1,Peptide from Integrin beta-1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
1R2X
 
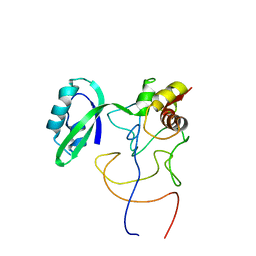 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
7SNA
 
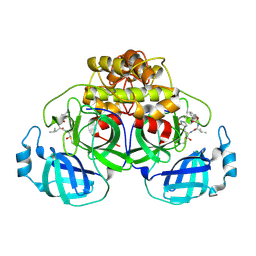 | | Crystallization of feline coronavirus Mpro with GC376 reveals mechanism of inhibition | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallization of Feline Coronavirus M pro With GC376 Reveals Mechanism of Inhibition.
Front Chem, 10, 2022
|
|
8GS2
 
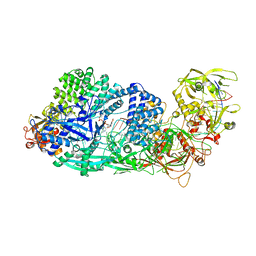 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
8GS5
 
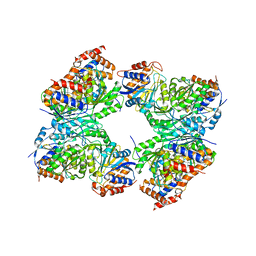 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRD
 
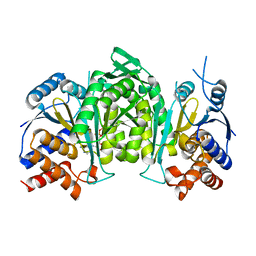 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRG
 
 | |
