2X0A
 
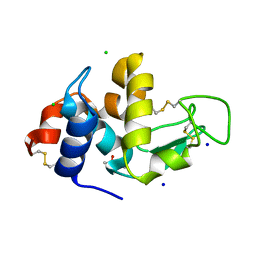 | |
2WJY
 
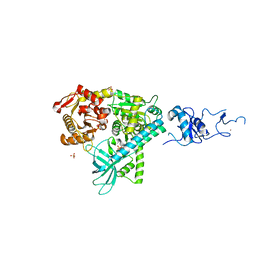 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 Orthorhombic form | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, SULFATE ION, ZINC ION | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
7QIQ
 
 | | CRYSTAL STRUCTURE OF THE P1 aminobutanoic acid (ABU) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
6H9N
 
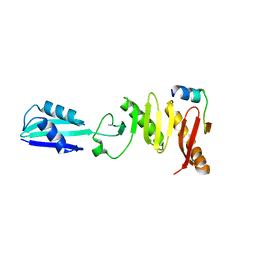 | |
3ZMM
 
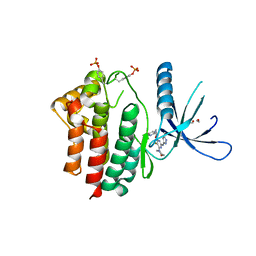 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7QQY
 
 | | yeast Gid10 bound to Art2 Pro/N-degron | | Descriptor: | CHLORIDE ION, ECM21, Uncharacterized protein YGR066C | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A GID E3 ligase assembly ubiquitinates an Rsp5 E3 adaptor and regulates plasma membrane transporters.
Embo Rep., 23, 2022
|
|
7QIR
 
 | | CRYSTAL STRUCTURE OF THE P1 monofluorethylglycine(MfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIS
 
 | | CRYSTAL STRUCTURE OF THE P1 difluoroethylglycine (DfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
7QIT
 
 | | CRYSTAL STRUCTURE OF THE P1 trifluoroethylglycine (TfeGly) BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Wahl, M.C, Loll, B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fluorine-induced polarity increases inhibitory activity of BPTI towards chymotrypsin.
Rsc Chem Biol, 3, 2022
|
|
6HJY
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) Delta8 truncation mutant in complex with nanobody 72 | | Descriptor: | Cys-loop ligand-gated ion channel, nanobody 72 | | Authors: | Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
7QCM
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3-methoxy-4-hydroxy-acetophenone)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-metoxy-4-hydroxy-acetophenone)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCK
 
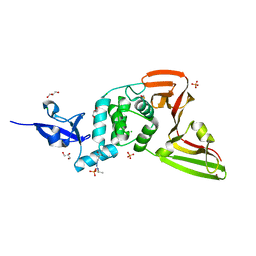 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,5-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,5-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QCI
 
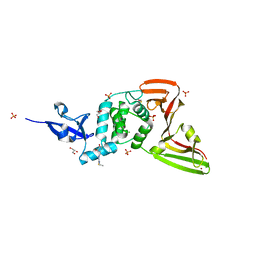 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
2VI1
 
 | |
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
4PT2
 
 | |
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
6HHJ
 
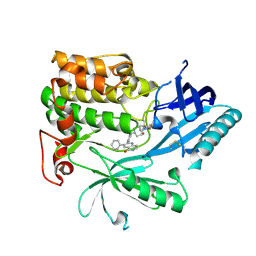 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 24b | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[1-methyl-2-oxidanylidene-3-[1-[[4-(5-oxidanylidene-3-phenyl-6~{H}-1,6-naphthyridin-2-yl)phenyl]methyl]piperidin-4-yl]benzimidazol-5-yl]propanamide | | Authors: | Landel, I, Weisner, J, Mueller, M.P, Scheinpflug, R, Rauh, D. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and chemical insights into the covalent-allosteric inhibition of the protein kinase Akt.
Chem Sci, 10, 2019
|
|
3ZVB
 
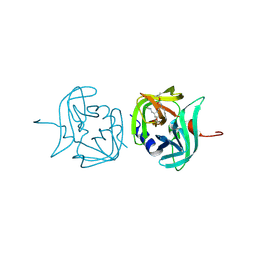 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 81 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
7QCG
 
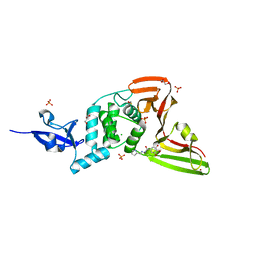 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
3NAP
 
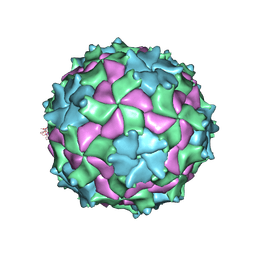 | | Structure of Triatoma Virus (TrV) | | Descriptor: | Capsid protein | | Authors: | Squires, G, Pous, J. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of Triatoma Virus (TrV) highlights the Dicistroviridae and Picornaviridae family differences
To be Published
|
|
3ZV8
 
 | |
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | Descriptor: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
5N89
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE GNSFDDWLASKG | | Descriptor: | GLY-ASN-SER-PHE-ASP-ASP-TRP-LEU-ALA-SER-LYS-GLY-NH2, GLYCEROL, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
