3I1E
 
 | | Crystal Structure of the PDZ domain of the SdrC-like Protein (Lin2157) from Listeria innocua, Northeast Structural Genomics Consortium Target LkR136C | | Descriptor: | Lin2157 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR136C
To be Published
|
|
4KV9
 
 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
3ZJJ
 
 | | Phe(93)E11Leu mutation of M.acetivorans protoglobin in complex with cyanide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYANIDE ION, GLYCEROL, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
3ZJS
 
 | | M.acetivorans protoglobin in complex with azide and Xenon | | Descriptor: | AZIDE ION, PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
3ZOQ
 
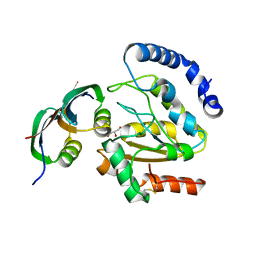 | | Structure of BsUDG-p56 complex | | Descriptor: | CHLORIDE ION, GLYCEROL, P56, ... | | Authors: | Banos-Sanz, J.I, Mojardin, L, Sanz-Aparicio, J, Gonzalez, B, Salas, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure and Functional Insights Into Uracil-DNA Glycosylase Inhibition by Phage Phi29 DNA Mimic Protein P56
Nucleic Acids Res., 41, 2013
|
|
3HXK
 
 | | Crystal Structure of a sugar hydrolase (YeeB) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR108 | | Descriptor: | Sugar hydrolase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-20 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a sugar hydrolase (YeeB) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR108
To be Published
|
|
3ZRW
 
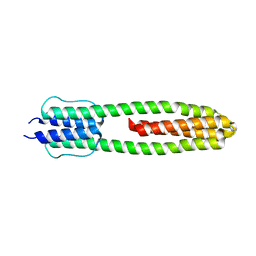 | | The structure of the dimeric Hamp-Dhp fusion A291V mutant | | Descriptor: | AF1503 PROTEIN, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J, Lupas, A.N. | | Deposit date: | 2011-06-20 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
3HNM
 
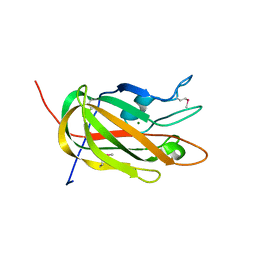 | | Crystal Structure of Protein BT_411 (putative chitobiase, fragment 298-461) from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR319D | | Descriptor: | MAGNESIUM ION, Putative chitobiase | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-31 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target BtR319D
To be published
|
|
3ZMR
 
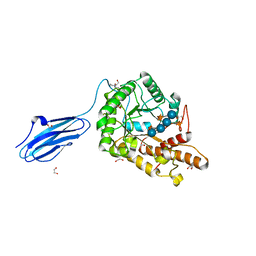 | | Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CELLULASE (GLYCOSYL HYDROLASE FAMILY 5), ... | | Authors: | Larsbrink, J, Rogers, T.E, Hemsworth, G.R, McKee, L.S, Spadiut, O, Klinter, S, Pudlo, N.A, Urs, K, Kelly, A.G, Cederholm, S.N, Davies, G.J, Martens, E.C, Brumer, H. | | Deposit date: | 2013-02-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Discrete Genetic Locus Confers Xyloglucan Metabolism in Select Human Gut Bacteroidetes
Nature, 506, 2014
|
|
3HQC
 
 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
1O93
 
 | | Methionine Adenosyltransferase complexed with ATP and a L-methionine analogue | | Descriptor: | (2S,4S)-2-AMINO-4,5-EPOXIPENTANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 2002-12-10 | | Release date: | 2003-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structures of Methionine Adenosyltransferase Complexed with Substrates and Products Reveal the Methionine-ATP Recognition and Give Insights Into the Catalytic Mechanism
J.Mol.Biol., 331, 2003
|
|
1Q7S
 
 | | Crystal structure of bit1 | | Descriptor: | bit1 | | Authors: | De Pereda, J.M, Waas, W.F, Jan, Y, Ruoslahti, E, Schimmel, P, Pascual, J. | | Deposit date: | 2003-08-19 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity.
J.Biol.Chem., 279, 2004
|
|
3ZDA
 
 | | Structure of E. coli ExoIX in complex with a fragment of the Flap1 DNA oligonucleotide, potassium and magnesium | | Descriptor: | 5'-D(*AP*AP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP)-3', MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZID
 
 | |
3HTJ
 
 | |
1Q8O
 
 | | Pterocartpus angolensis lectin PAL in complex with the dimmanoside Man(alpha1-2)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3ZL1
 
 | | A thiazolyl-mannoside bound to FimH, monoclinic space group | | Descriptor: | CHLORIDE ION, N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | Authors: | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
3K0W
 
 | | Crystal structure of the tandem IG-like C2-type 2 domains of the human mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Descriptor: | CHLORIDE ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, isoform 2 | | Authors: | Walker, J.R, Qiu, L, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tandem Ig-Like C2-Type 2 Domains of the Human Mucosa-Associated Lymphoid Tissue Lymphoma Translocation Protein 1.
To be Published
|
|
3ZN3
 
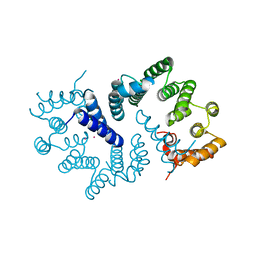 | | N-terminal domain of S. pombe Cdc23 APC subunit | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8, MERCURY (II) ION | | Authors: | Zhang, Z, Yang, J, Conin, N, Kulkarni, K, Barford, D. | | Deposit date: | 2013-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Four Canonical Tpr Subunits of Human Apc/C Form Related Homo-Dimeric Structures and Stack in Parallel to Form a Tpr Suprahelix
J.Mol.Biol., 425, 2013
|
|
4NB2
 
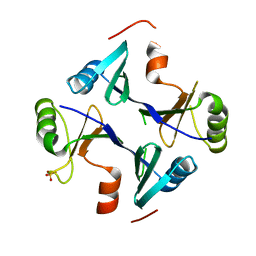 | | Crystal Structure of FosB from Staphylococcus aureus at 1.89 Angstrom Resolution - Apo structure | | Descriptor: | Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
3JTK
 
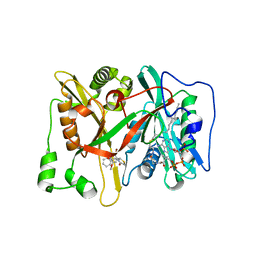 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055 | | Descriptor: | (2R)-3-benzyl-2-(2-bromo-4-hydroxy-5-methoxyphenyl)-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90055
To be Published
|
|
3ZQ1
 
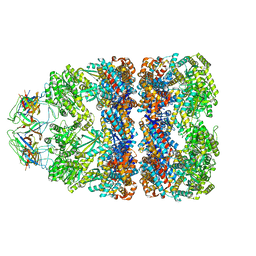 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
3K1Z
 
 | | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Haloacid dehalogenase-like hydrolase domain-containing protein 3 | | Authors: | Ugochukwu, E, Guo, K, Yue, W.W, Pilka, E, Picaud, S, Muniz, J, Pike, A.C.W, Krojer, T, Gomes, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-11-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3)
To be Published
|
|
3K25
 
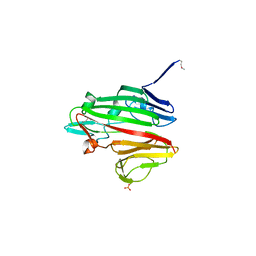 | | Crystal Structure of Slr1438 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR112 | | Descriptor: | PHOSPHATE ION, Slr1438 protein | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR112
To be published
|
|
