1IPE
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
2RGW
 
 | |
2RFT
 
 | | Crystal structure of influenza B virus hemagglutinin in complex with LSTa receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Influenza B hemagglutinin (HA), ... | | Authors: | Wang, Q, Tian, X, Chen, X, Ma, J. | | Deposit date: | 2007-10-01 | | Release date: | 2008-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor specificity of influenza B virus hemagglutinin.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | Descriptor: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | Authors: | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
5KQU
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
7E2R
 
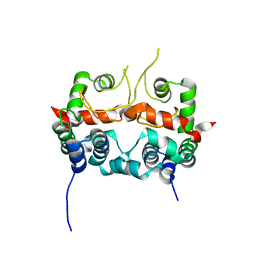 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
5KRM
 
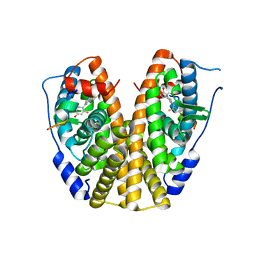 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2,5-difluoro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | Descriptor: | (1~{S},3~{a}~{R},7~{a}~{S})-5-[2,5-bis(fluoranyl)-4-oxidanyl-phenyl]-7~{a}-methyl-1,2,3,3~{a},4,7-hexahydroinden-1-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6IP4
 
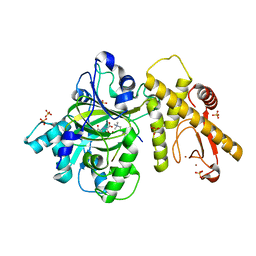 | |
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
2RIR
 
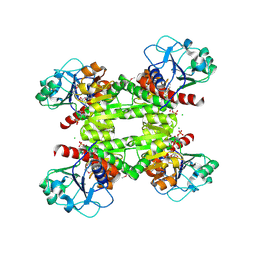 | | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis | | Descriptor: | CHLORIDE ION, Dipicolinate synthase, A chain, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis.
To be Published
|
|
2RMM
 
 | |
2RO3
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator Abh | | Descriptor: | Putative transition state regulator abh | | Authors: | Sullivan, D.M, Bobay, B.G, Douglas, K.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
5KQL
 
 | |
2RM5
 
 | | Glutathione peroxidase-type tryparedoxin peroxidase, oxidized form | | Descriptor: | Glutathione peroxidase-like protein | | Authors: | Melchers, J, Feher, K, Diechtierow, M, Krauth-Siegel, L, Tews, I, Muhle-Goll, C. | | Deposit date: | 2007-10-03 | | Release date: | 2008-07-29 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a distinct catalytic mechanism in Trypanosoma brucei tryparedoxin peroxidase
J.Biol.Chem., 283, 2008
|
|
5KQP
 
 | | Crystal structure of Apo-form LMW-PTP | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Wang, J, Zhang, Z.-Y, Yu, Z.-H. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Inhibition of low molecular weight protein tyrosine phosphatase by an induced-fit mechanism.
J.Med.Chem., 2016
|
|
5KR9
 
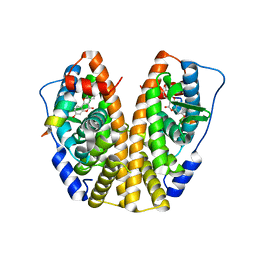 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Coumestrol | | Descriptor: | Coumestrol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2R5H
 
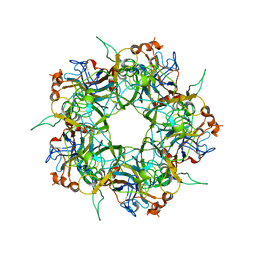 | |
5KRK
 
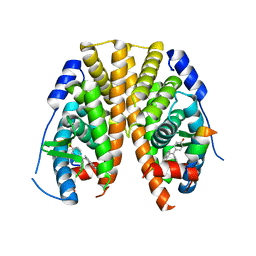 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-((5-bromo-2,3-dihydro-1H-inden-1-ylidene)methylene)diphenol | | Descriptor: | 4-[(5-bromanyl-2,3-dihydroinden-1-ylidene)-(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2RBG
 
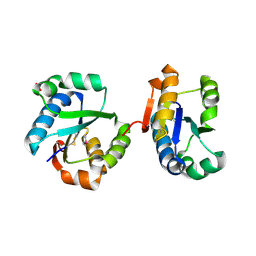 | |
5KYY
 
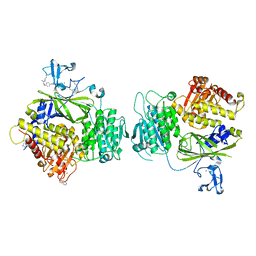 | | Crystal structure of Sec23 and TANGO1 peptide4 complex | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24D, ZINC ION | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2016-07-22 | | Release date: | 2016-09-07 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2R5J
 
 | |
6ISB
 
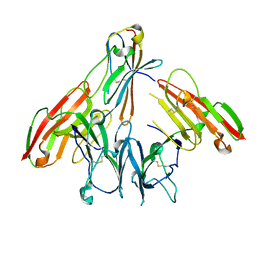 | | crystal structure of human CD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2R8Q
 
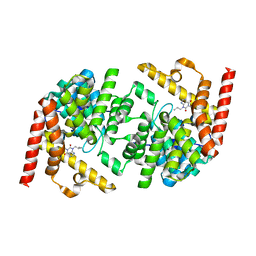 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
5KYX
 
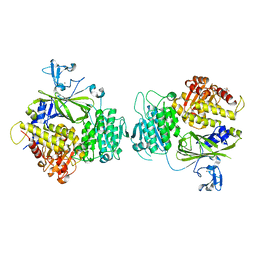 | | crystal structure of Sec23 and TANGO1 peptide1 complex | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24D, ZINC ION | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2016-07-22 | | Release date: | 2016-09-07 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RIB
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with L-glycero-D-manno-heptose | | Descriptor: | CALCIUM ION, L-glycero-alpha-D-manno-heptopyranose, Pulmonary surfactant-associated protein D | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
