6GOO
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with Perfluorooctanoic acid (PFOA). Crystallized in AmSO4 | | Descriptor: | Transthyretin, pentadecafluorooctanoic acid | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
8TSM
 
 | |
1YAT
 
 | |
4DLO
 
 | | Crystal structure of the GAIN and HormR domains of brain angiogenesis inhibitor 3 (BAI3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brain-specific angiogenesis inhibitor 3, ... | | Authors: | Arac, D, Boucard, A.A, Bolliger, M.F, Nguyen, J, Soltis, M, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis.
Embo J., 31, 2012
|
|
6GQM
 
 | | Crystal structure of human c-KIT kinase domain in complex with a small molecule inhibitor, AZD3229 | | Descriptor: | CHLORIDE ION, Mast/stem cell growth factor receptor Kit, ~{N}-[4-[[5-fluoranyl-7-(2-methoxyethoxy)quinazolin-4-yl]amino]phenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
4L3B
 
 | | X-ray structure of the HRV2 A particle uncoating intermediate | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Vives-Adrian, L, Querol-Audi, J, Garriga, D, Pous, J, Verdaguer, N. | | Deposit date: | 2013-06-05 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2VA5
 
 | | X-ray crystal structure of beta secretase complexed with compound 8c | | Descriptor: | 2-amino-6-[2-(1H-indol-6-yl)ethyl]pyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
2O9R
 
 | | beta-glucosidase B complexed with thiocellobiose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
2VEY
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[4-(3-phenylpropyl)-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2OBK
 
 | | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens. Northeast Structural Genomics Consortium target PlR6. | | Descriptor: | SelT/selW/selH selenoprotein domain | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens
To be Published
|
|
1CON
 
 | | THE REFINED STRUCTURE OF CADMIUM SUBSTITUTED CONCANAVALIN A AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CADMIUM ION, CALCIUM ION, CONCANAVALIN A | | Authors: | Naismith, J.H, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Kalb(Gilboa), A.J, Yariv, J, Wan, T.C.M, Weisgerber, S. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2VCO
 
 | | Crystal structure of the fimbrial adhesin FimH in complex with its high-mannose epitope | | Descriptor: | NICKEL (II) ION, PROTEIN FIMH, SULFATE ION, ... | | Authors: | Wellens, A, Garofalo, C, Nguyen, H, Wyns, L, De Greve, H, Hultgren, S.J, Bouckaert, J. | | Deposit date: | 2007-09-26 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the Fimh-Oligomannose-3 Complex.
Plos One, 3, 2008
|
|
8TXQ
 
 | | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | CBH-4B Heavy chain, CBH-4B Light chain, envelope glycoprotein E2 | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of non-neutralizing antibody CBH-4B in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
2O8S
 
 | | X-ray Crystal Structure of Protein AGR_C_984 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR120. | | Descriptor: | AGR_C_984p, SULFATE ION | | Authors: | Seetharaman, J, Forouhar, F, Su, M, Zhao, L, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Hypothetical Protein AGR_C_984 from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR120.
To be Published
|
|
8TK6
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) in apo form | | Descriptor: | Dual specificity protein phosphatase 3 | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment Screening Platform and Discovery of Novel Fragment Binders of the VHR Phosphatase, a Drug Target for Sepsis and Septic Shock
To Be Published
|
|
2VAT
 
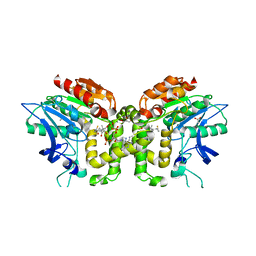 | | Crystal structure of deacetylcephalosporin C acetyltransferase in complex with coenzyme A | | Descriptor: | ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE, COENZYME A, ... | | Authors: | Lejon, S, Ellis, J, Valegard, K. | | Deposit date: | 2007-09-04 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
2VHI
 
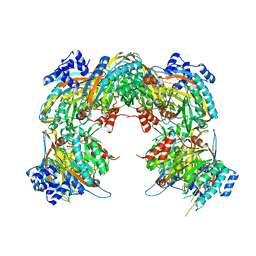 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
6GZR
 
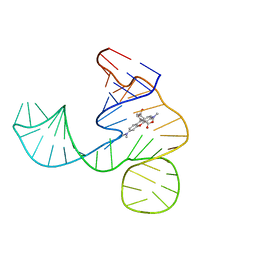 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
7DMW
 
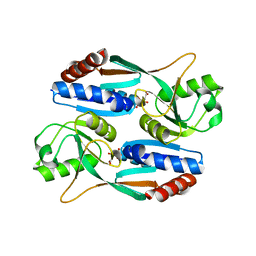 | | Crystal structure of CcpC regulatory domain in complex with citrate from Bacillus amyloliquefaciens | | Descriptor: | CITRATE ANION, CcpC | | Authors: | Chen, J, Wang, L, Shang, F, Liu, W, Chen, Y, Lan, J, Bu, T, Bai, X, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural analysis of catabolite control protein C that responds to citrate.
Sci Rep, 11, 2021
|
|
8TK4
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with phosphate | | Descriptor: | Dual specificity protein phosphatase 3, PHOSPHATE ION | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with phosphate
To Be Published
|
|
8TON
 
 | |
8TS3
 
 | | Cryo-EM structure of human MRS2 with Ca2+ | | Descriptor: | CALCIUM ION, Magnesium transporter MRS2 homolog, mitochondrial, ... | | Authors: | He, Z, Mount, J.W, Zhang, J, Yuan, P. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure and function of the human mitochondrial MRS2 channel.
Nat.Struct.Mol.Biol., 2024
|
|
8TK5
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE (VHR) complexed with HEPES | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aleshin, A.E, Wu, J, Lambert, L.J, Cosford, N.D.P, Tautz, L. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment Screening Platform and Discovery of Novel Fragment Binders of the VHR Phosphatase, a Drug Target for Sepsis and Septic Shock
To Be Published
|
|
8TS2
 
 | | Cryo-EM structure of human MRS2 with EDTA | | Descriptor: | Magnesium transporter MRS2 homolog, mitochondrial, Soluble cytochrome b562 fusion protein | | Authors: | He, Z, Mount, J.W, Zhang, J, Yuan, P. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure and function of the human mitochondrial MRS2 channel.
Nat.Struct.Mol.Biol., 2024
|
|
4NFU
 
 | | Structure of the central plant immunity signaling node EDS1 in complex with its interaction partner SAG101 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, EDS1, ISOPROPYL ALCOHOL, ... | | Authors: | Wagner, S, Stuttmann, J, Rietz, S, Guerois, R, Niefind, K, Parker, J.E. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity.
Cell Host Microbe, 14, 2013
|
|
