3LDW
 
 | | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Wernimont, A.K, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
2YGW
 
 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
6A7H
 
 | | Bacterial protein toxins | | Descriptor: | RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
2ZOX
 
 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
2OWA
 
 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
4P0U
 
 | |
2JSA
 
 | |
4NHW
 
 | | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275, with one glutathione bound per one protein subunit | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275
To be Published
|
|
2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
2P65
 
 | | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax | | Descriptor: | Hypothetical protein PF08_0063 | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax
To be Published
|
|
2P6E
 
 | |
3QKI
 
 | | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7 | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Gileadi, T, Wernimont, A.K, Hutchinson, A, Weadge, J, Cossar, D, Lew, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Glucose-6-Phosphate Isomerase (PF14_0341) from Plasmodium falciparum 3D7
TO BE PUBLISHED
|
|
4NS0
 
 | | The C2A domain of Rabphilin 3A in complex with PI(4,5)P2 | | Descriptor: | Rabphilin-3A, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2ONU
 
 | | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H | | Descriptor: | TETRAETHYLENE GLYCOL, Ubiquitin-conjugating enzyme, putative | | Authors: | Dong, A, Lew, J, Lin, L, Hassanali, A, Zhao, Y, Senisterra, G, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Hui, R, Brokx, S.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Plasmodium falciparum ubiquitin conjugating enzyme PF10_0330, putative homologue of human UBE2H
To be Published
|
|
2OO2
 
 | | Crystal structure of protein AF1782 from Archaeoglobus fulgidus, Pfam DUF357 | | Descriptor: | Hypothetical protein AF_1782 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Adams, J, Sridhar, V, Smyth, L, Freeman, J, Atwell, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical AF_1782 protein from Archaeoglobus fulgidus
To be Published
|
|
4NCA
 
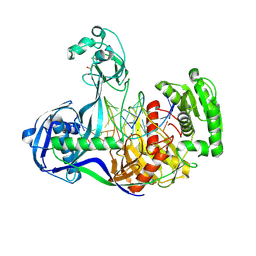 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | Descriptor: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-24 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OPG
 
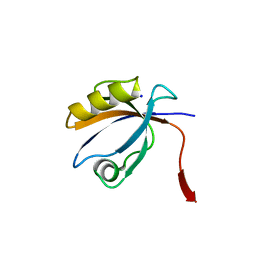 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
6AJH
 
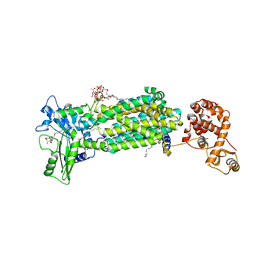 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
2JF6
 
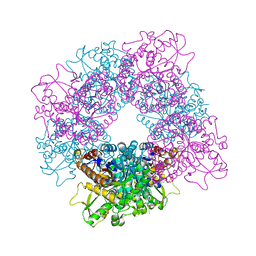 | | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine | | Descriptor: | METHYL (2S,3R,4S)-3-ETHYL-2-(BETA-D-GLUCOPYRANOSYLOXY)-4-[(1S)-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLIN-1-YLMETHYL]-3,4-DIHYDRO-2H-PYRAN-5-CARBOXYLATE, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Barleben, L, Panjikar, S, Ruppert, M, Koepke, J, Stockigt, J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Molecular Architecture of Strictosidine Glucosidase - the Gateway to the Biosynthesis of the Monoterpenoid Indole Alkaloid Family
Plant Cell, 19, 2007
|
|
2JIE
 
 | |
