5FSM
 
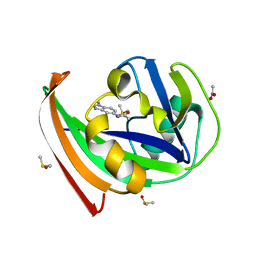 | | MTH1 substrate recognition: Complex with a methylbenzimidazolyl acetamide. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5FN0
 
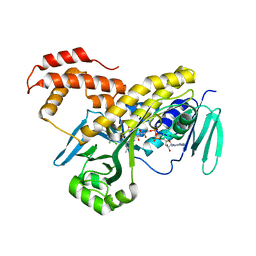 | | Crystal structure of Pseudomonas fluorescens kynurenine-3- monooxygenase (KMO) in complex with GSK180 | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, KYNURENINE 3-MONOOXYGENASE | | Authors: | Mole, D.J, Webster, S.P, Uings, I, Zheng, X, Binnie, M, Wilson, K, Hutchinson, J.P, Mirguet, O, Walker, A, Beaufils, B, Ancellin, N, Trottet, L, Beneton, V, Mowat, C.G, Wilkinson, M, Rowland, P, Haslam, C, McBride, A, Homer, N.Z.M, Baily, J.E, Sharp, M.G.F, Garden, O.J, Hughes, J, Howie, S.E.M, Holmes, D, Liddle, J, Iredale, J.P. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Kynurenine-3-Monooxygenase Inhibition Prevents Multiple Organ Failure in Rodent Models of Acute Pancreatitis.
Nat.Med. (N.Y.), 22, 2016
|
|
6OC5
 
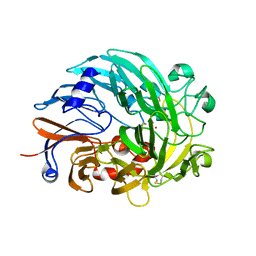 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
5FSN
 
 | | MTH1 substrate recognition: Complex with a aminomethylpyrimidinyl oxypropanol. | | Descriptor: | 3-(2-AMINO-6-METHYL-PYRIMIDIN-4-YL)OXYPROPAN-1-OL, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5FSK
 
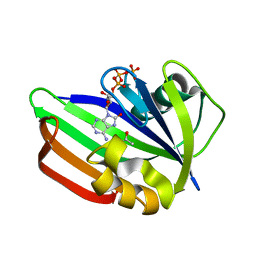 | | MTH1 substrate recognition: Complex with 8-oxo-dGTP. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-ADENOSINE-5'-TRIPHOSPHATE, ACETATE ION | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
4FPR
 
 | | Structure of a fungal protein | | Descriptor: | Avirulence Effector AvrLm4-7 | | Authors: | Blondeau, K, Blaise, F, Graille, M, Linglin, J, Ollivier, B, Labarde, A, Doizy, A, Daverdin, G, Balesdent, M.H, Rouxel, T, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2012-06-22 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of the effector AvrLm4-7 of Leptosphaeria maculans reveals insights into its translocation into plant cells and recognition by resistance proteins.
Plant J., 83, 2015
|
|
6OSQ
 
 | | RF1 accommodated state bound Release complex 70S at long incubation time point | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-26 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
6OL2
 
 | | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high-throughput-screening | | Descriptor: | ACETATE ION, GLYCEROL, N-[2-(5,8-dimethoxy-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]-2-iodobenzamide, ... | | Authors: | Chlebowicz, J, Akella, R, Sekulski, K, Humphreys, J.M, Durbacz, M.Z, He, H, Rodan, A, Posner, B, Goldsmith, E.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high throughput screening
To Be Published
|
|
5FT4
 
 | | Crystal structure of the cysteine desulfurase CsdA from Escherichia coli at 1.996 Angstroem resolution | | Descriptor: | CITRIC ACID, CYSTEINE DESULFURASE CSDA, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-12-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | The Mechanism of Sulfur Transfer Across Protein- Protein Interfaces: The Csd Model
Acs Catalysis, 6, 2016
|
|
2LLV
 
 | | Solution structure of the yeast Sti1 DP1 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
2L6X
 
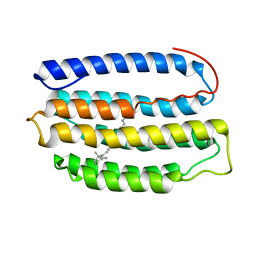 | | Solution NMR Structure of Proteorhodopsin. | | Descriptor: | Green-light absorbing proteorhodopsin, RETINAL | | Authors: | Reckel, S, Gottstein, D, Stehle, J, Loehr, F, Takeda, M, Silvers, R, Kainosho, M, Glaubitz, C, Bernhard, F, Schwalbe, H, Guntert, P, Doetsch, V, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of proteorhodopsin.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6OC6
 
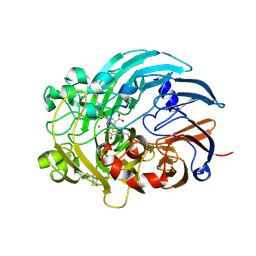 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum and Pyrroloquinoline quinone | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
6OTW
 
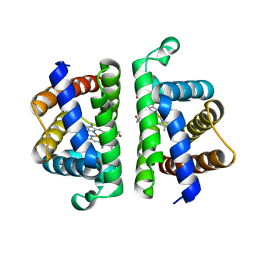 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
6OTY
 
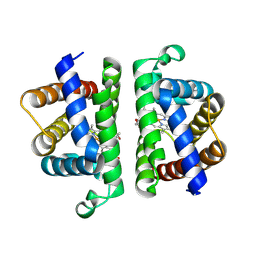 | | Crystallographic Structure of (HbII-HbIII)-O2 from Lucina pectinata at pH 4.0 | | Descriptor: | Hemoglobin II, Hemoglobin III, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Marchany-Rivera, D, Smith, C.A, Rodriguez-Perez, J.D, Lopez-Garriga, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Lucina pectinata oxyhemoglobin (II-III) heterodimer pH susceptibility.
J.Inorg.Biochem., 207, 2020
|
|
7XVK
 
 | |
2L89
 
 | |
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7XVI
 
 | |
4FMK
 
 | | Crystal structure of mouse nectin-2 extracellular fragment D1-D2 | | Descriptor: | CADMIUM ION, Poliovirus receptor-related protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-17 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6OF9
 
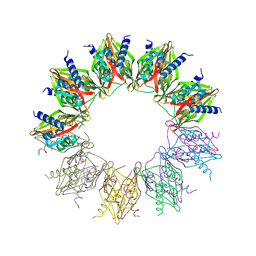 | |
2LEC
 
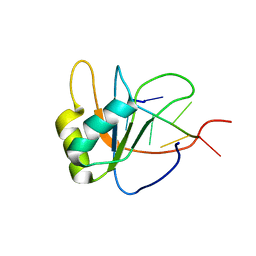 | | Solution structure of human SRSF2 (SC35) RRM in complex with 5'-UGGAGU-3' | | Descriptor: | RNA (5'-R(*UP*GP*GP*AP*GP*U)-3'), Serine/arginine-rich splicing factor 2 | | Authors: | Daubner, G.M, Clery, A, Jayne, S, Stevenin, J, Allain, F.H.-T. | | Deposit date: | 2011-06-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Embo J., 31, 2012
|
|
6OIG
 
 | | Subunit joining exposes nascent pre-40S rRNA for processing and quality control | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Rai, J, Parker, M.D, Ghalei, H, Johnson, M.C, Karbstein, K, Stroupe, M.E. | | Deposit date: | 2019-04-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An open interface in the pre-80S ribosome coordinated by ribosome assembly factors Tsr1 and Dim1 enables temporal regulation of Fap7.
Rna, 27, 2021
|
|
4FQP
 
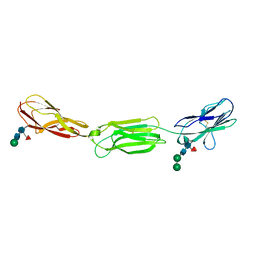 | | Crystal structure of human Nectin-like 5 full ectodomain (D1-D3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, ... | | Authors: | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6OEU
 
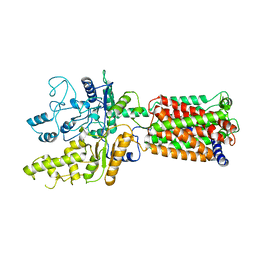 | | Structure of human Patched1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Qi, X, Li, X, Wang, J. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
7RF8
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
