5LPZ
 
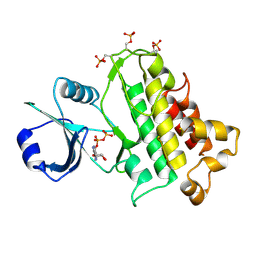 | |
8D56
 
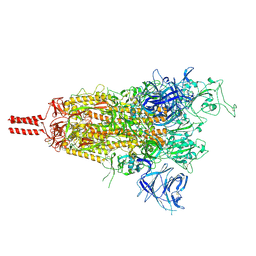 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6J65
 
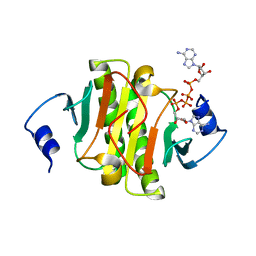 | | Crystal structure of human HINT1 mutant complexing with AP4A II | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8D55
 
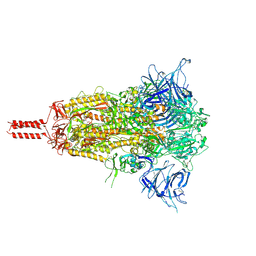 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
8GJW
 
 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
8DIR
 
 | |
2CA7
 
 | | Conkunitzin-S1 Is The First Member Of A New Kunitz-Type Neurotoxin Family- Structural and Functional Characterization | | Descriptor: | CONKUNITZIN-S1 | | Authors: | Bayrhuber, M, Vijayan, V, Ferber, M, Graf, R, Korukottu, J, Imperial, J, Garrett, J.E, Olivera, B.M, Terlau, H, Zweckstetter, M, Becker, S. | | Deposit date: | 2005-12-19 | | Release date: | 2006-01-05 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization.
J. Biol. Chem., 280, 2005
|
|
4Y3F
 
 | |
8D9Y
 
 | | Crystal structure of Taipan alpha-neurotoxin in complex with Centi-3FTX-D09 antibody | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Centi-3FTX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
5MLL
 
 | |
8CEV
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO1119 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-iodanyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
4Y5L
 
 | | Endothiapepsin in its apo form | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
8CER
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO494 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-naphthalen-1-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
8CEQ
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO427 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-phenylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8CES
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO500 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-[2-(1~{H}-benzimidazol-2-yl)ethynyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
6W22
 
 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8CET
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO507 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
6ZZZ
 
 | |
6ZPR
 
 | | Solution structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein,Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-06 | | Method: | SOLUTION NMR | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GL9
 
 | | Co-crystal structure of caPCNA bound to AOH1160 derivative 1LE | | Descriptor: | CHLORIDE ION, N~2~-(naphthalene-1-carbonyl)-N-(2-phenoxyphenyl)-L-alpha-glutamine, Proliferating cell nuclear antigen | | Authors: | Jossart, J, Kenjic, N, Malkas, L.H, Hickey, R.J, Perry, J.J. | | Deposit date: | 2023-03-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Small molecule targeting of transcription-replication conflict for selective chemotherapy.
Cell Chem Biol, 30, 2023
|
|
8GLA
 
 | |
