5GSV
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
6HCU
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum bound to a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | Authors: | Tamjar, J, Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5GSR
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
4OC4
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CPIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(pyridin-4-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5GR7
 
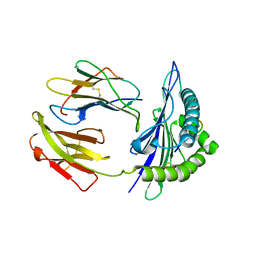 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
4A1X
 
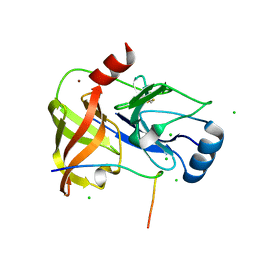 | | Co-Complex structure of NS3-4A protease with the inhibitory peptide CP5-46-A (Synchrotron data) | | Descriptor: | CHLORIDE ION, CP5-46-A PEPTIDE, NONSTRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4A1V
 
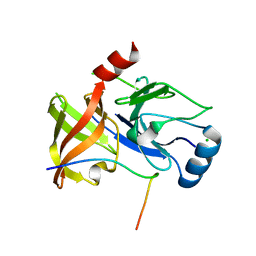 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
5GSB
 
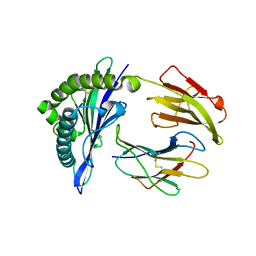 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
4RLP
 
 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5H1V
 
 | | Complex structure of TRIM24 PHD-bromodomain and inhibitor 6 | | Descriptor: | 2-Hydrazino-1,3-benzothiazole-6-carbohydrazide, DIMETHYL SULFOXIDE, Transcription intermediary factor 1-alpha, ... | | Authors: | Liu, J. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The polar warhead of a TRIM24 bromodomain inhibitor rearranges a water-mediated interaction network
FEBS J., 284, 2017
|
|
2WD1
 
 | | Human c-Met Kinase in complex with azaindole inhibitor | | Descriptor: | 1-[(2-NITROPHENYL)SULFONYL]-1H-PYRROLO[3,2-B]PYRIDINE-6-CARBOXAMIDE, GAMMA-BUTYROLACTONE, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Porter, J, Lumb, S, Franklin, R.J, Gascon-Simorte, J.M, Calmiano, M, Le Riche, K, Lallemand, B, Keyaerts, J, Edwards, H, Maloney, A, Delgado, J, King, L, Foley, A, Lecomte, F, Reuberson, J, Meier, C, Batchelor, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-Azaindoles as Novel Inhibitors of C- met Kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2XIN
 
 | |
4RLO
 
 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
2ERM
 
 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
4PVV
 
 | | Micobacterial Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase | | Authors: | Pichova, I, Hocek, M, Dostal, J, Rezacova, P. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
2F6I
 
 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
2F5J
 
 | | Crystal structure of MRG domain from human MRG15 | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Zhang, P, Du, J, Ding, J. | | Deposit date: | 2005-11-26 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
Protein Sci., 15, 2006
|
|
1Y6Z
 
 | | Middle domain of Plasmodium falciparum putative heat shock protein PF14_0417 | | Descriptor: | heat shock protein, putative | | Authors: | Lunin, V.V, Botchkareva, E, Loppnau, P, Amani, M, Bray, J, Vedadi, M, Edwards, A, Arrowsmith, C, Sundstrom, M, Bochkarev, A, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-07 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2DA8
 
 | | SOLUTION STRUCTURE OF A COMPLEX BETWEEN (N-MECYS3,N-MECYS7)TANDEM AND (D(GATATC))2 | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*GP*AP*TP*AP*TP*C)-3'), TRIOSTIN A | | Authors: | Addess, K.J, Sinsheimer, J.S, Feigon, J. | | Deposit date: | 1993-04-09 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Complex between [N-Mecys3,N-Mecys7]Tandem and [D(Gatatc)]2.
Biochemistry, 32, 1993
|
|
1TKZ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW429576 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFANYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TKX
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
2AAS
 
 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | RIBONUCLEASE A | | Authors: | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | Deposit date: | 1992-11-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
1TL3
 
 | | Crystal structure of hiv-1 reverse transcriptase in complex with gw450557 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-ISOPROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
6DHF
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
