4CN3
 
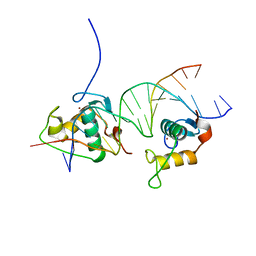 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human Gde1SpA Response Element | | Descriptor: | 5'-D(*CP*TP*AP*GP*TP*TP*CP*AP*AP*AP*GP*TP*TP*CP *AP*CP*A)-3', 5'-D(*TP*GP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*AP*CP *TP*AP*G)-3', RETINOIC ACID RECEPTOR RXR-ALPHA, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2014-01-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Natural Promoter Recognition by the Retinoid X Nuclear Receptor.
Sci.Rep., 5, 2015
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-12-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
5PRM
 
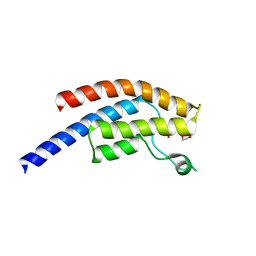 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 94) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6HGS
 
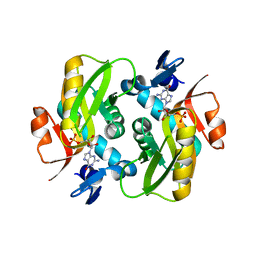 | | Crystal Structure of Human APRT wild type in complex with GMP | | Descriptor: | Adenine phosphoribosyltransferase, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for substrate selectivity and nucleophilic substitution mechanisms in human adenine phosphoribosyltransferase catalyzed reaction.
J.Biol.Chem., 294, 2019
|
|
5PS3
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 110) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4CV1
 
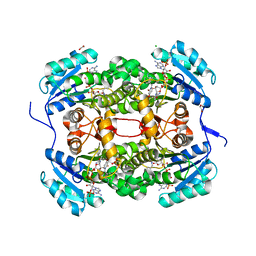 | | Crystal structure of S. aureus FabI in complex with NADPH and CG400549 | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-amino-2-methylbenzyl)-4-[2-(thiophen-2-yl)ethoxy]pyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Schiebel, J, Chang, A, Shah, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
5PLX
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 155) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4CXL
 
 | | Human insulin analogue (D-ProB8)-insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
6BP8
 
 | | Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 1 | | Descriptor: | Major vault protein | | Authors: | Ding, K, Zhang, X, Mrazek, J, Kickhoefer, V.A, Lai, M, Ng, H.L, Yang, O.O, Rome, L.H, Zhou, Z.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Solution Structures of Engineered Vault Particles.
Structure, 26, 2018
|
|
5PM8
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 166) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PMN
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 181) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6HHC
 
 | | Allosteric Inhibition as a new mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated form of Coagulation Factor XI | | Descriptor: | Coagulation factor XI, DIMETHYL SULFOXIDE, FXIA ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Schaefer, M, Buchmueller, A, Dittmer, F, Strassburger, J, Wilmen, A. | | Deposit date: | 2018-08-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition as a New Mode of Action for BAY 1213790, a Neutralizing Antibody Targeting the Activated Form of Coagulation Factor XI.
J.Mol.Biol., 431, 2019
|
|
5PSF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 123) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSU
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 138) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PNE
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 208) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PNI
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D after initial refinement with no ligand modelled (structure 212) | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, MAGNESIUM ION, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3CMZ
 
 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
4D45
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-bromo- 2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
1FG0
 
 | | LARGE RIBOSOMAL SUBUNIT COMPLEXED WITH A 13 BP MINIHELIX-PUROMYCIN COMPOUND | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(CCGGCGGGCUGGUUCAAACCGGCCCGCCGGACC)-3'-5'-R(P-PUROMYCIN)-3' | | Authors: | Nissen, P, Hansen, J, Ban, N, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of ribosome activity in peptide bond synthesis.
Science, 289, 2000
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
6HUY
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstitued with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form A | | Descriptor: | 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, DIMETHYL SULFOXIDE, ... | | Authors: | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4CJ7
 
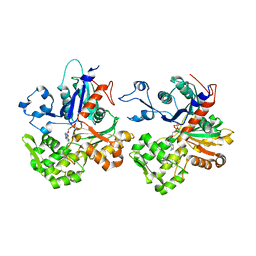 | | Structure of Crenactin, an archeal actin-like protein | | Descriptor: | ACTIN/ACTIN FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Izore, T, Duman, R.E, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crenactin from Pyrobaculum Calidifontis is Closely Related to Actin in Structure and Forms Steep Helical Filaments.
FEBS Lett., 588, 2014
|
|
5V10
 
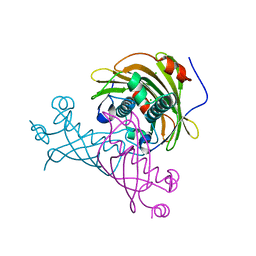 | | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Borek, D, Wawrzak, Z, Grimshaw, S, Sandoval, J, Evdokimova, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1
To Be Published
|
|
3CON
 
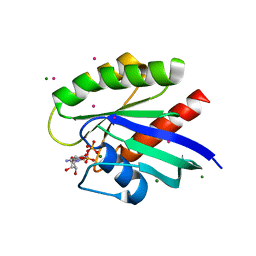 | | Crystal structure of the human NRAS GTPase bound with GDP | | Descriptor: | GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nedyalkova, L, Tong, Y, Tempel, W, Shen, L, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal structure of the human NRAS GTPase bound with GDP.
To be Published
|
|
6HWM
 
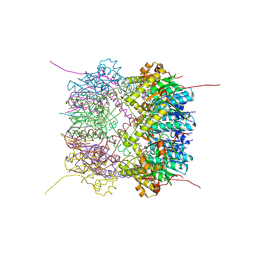 | | Structure of Thermus thermophilus ClpP in complex with bortezomib | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, DI(HYDROXYETHYL)ETHER, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Felix, J, Schanda, P, Fraga, H, Morlot, C. | | Deposit date: | 2018-10-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors.
Sci Adv, 5, 2019
|
|
