4TK0
 
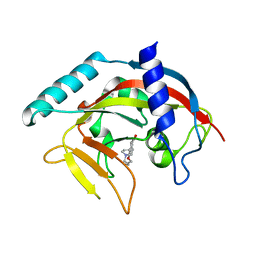 | | Crystal Structure of human Tankyrase 2 in complex with DPQ. | | Descriptor: | 5-[4-(piperidin-1-yl)butoxy]-3,4-dihydroisoquinolin-1(2H)-one, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JZZ
 
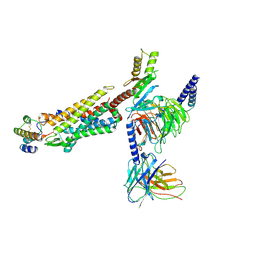 | | Structure of human C5a-desArg bound human C5aR1 in complex with Go | | Descriptor: | Antibody fragment ScFv16, C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-07-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
1CHL
 
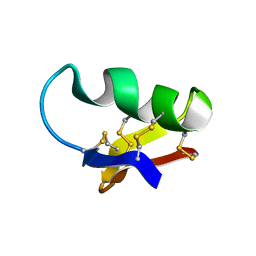 | | NMR SEQUENTIAL ASSIGNMENTS AND SOLUTION STRUCTURE OF CHLOROTOXIN, A SMALL SCORPION TOXIN THAT BLOCKS CHLORIDE CHANNELS | | Descriptor: | CHLOROTOXIN | | Authors: | Lippens, G, Najib, J, Wodak, S.J, Tartar, A. | | Deposit date: | 1994-11-09 | | Release date: | 1995-02-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels.
Biochemistry, 34, 1995
|
|
1YBK
 
 | | RHCC cocrystallized with CAPB | | Descriptor: | N-(CARBOXYMETHYL)-N,N-DIMETHYL-3-[(1-OXODODECYL)AMINO]-1-PROPANAMINIUM INNER SALT, SULFATE ION, tetrabrachion | | Authors: | Stetefeld, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Favourable mediation of crystal contacts by cocoamidopropylbetaine (CAPB).
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YI0
 
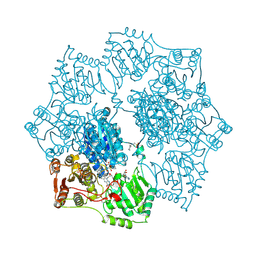 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Sulfometuron methyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ETHYL DIHYDROGEN DIPHOSPHATE, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5D4N
 
 | | Structure of CPII bound to ADP, AMP and acetate, from Thiomonas intermedia K12 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
4R3V
 
 | | Structure of karilysin propeptide and catalytic MMP domain | | Descriptor: | CALCIUM ION, GLYCEROL, Karilysin, ... | | Authors: | Lopez-Pelegrin, M, Ksiazek, M, Karim, A.Y, Guevara, T, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2014-08-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mechanism of latency in matrix metalloproteinases.
J.Biol.Chem., 290, 2015
|
|
4RBS
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, ACETIC ACID, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 in the Complex with Hydrolyzed Meropenem
To be Published
|
|
5D6O
 
 | | Orthorhombic Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | CHLORIDE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Niefind, K, Toelzer, C, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
5D14
 
 | |
1Y54
 
 | | Crystal structure of the native class C beta-lactamase from Enterobacter cloacae 908R complexed with BRL42715 | | Descriptor: | (7R)-6-FORMYL-7-(1-METHYL-1H-1,2,3-TRIAZOL-4-YL)-4,7-DIHYDRO-1,4-THIAZEPINE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Michaux, C, Charlier, P, Frere, J.-M, Wouters, J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of BRL 42715, C6-(N1-Methyl-1,2,3-triazolylmethylene)penem, in Complex with Enterobactercloacae 908R beta-Lactamase: Evidence for a Stereoselective Mechanism from Docking Studies
J.Am.Chem.Soc., 127, 2005
|
|
8K4R
 
 | | Structure of VinM-VinL complex | | Descriptor: | Acyl-carrier-protein, Non-ribosomal peptide synthetase, SODIUM ION, ... | | Authors: | Miyanaga, A, Nagata, K, Nakajima, J, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2023-07-20 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4RED
 
 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
3QKJ
 
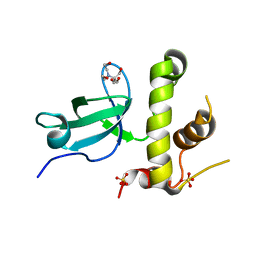 | | The PWWP domain of human DNA (CYTOSINE-5-)-METHYLTRANSFERASE 3 BETA in complex with a bis-tris molecule | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA cytosine-5 methyltransferase 3 beta isoform 6 variant, SULFATE ION | | Authors: | Zeng, H, Amaya, M.F, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
4R6L
 
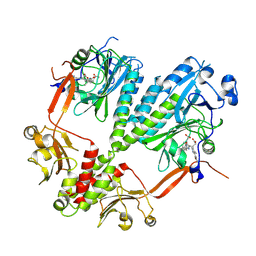 | | Crystal structure of bacteriophytochrome RpBphP2 from photosynthetic bacterium R. palustris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome (Light-regulated signal transduction histidine kinase), PhyB1 | | Authors: | Yang, X, Stojkovic, E, Ozarowski, W, Kuk, J, Davydova, E, Moffat, K. | | Deposit date: | 2014-08-25 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.395 Å) | | Cite: | Light Signaling Mechanism of Two Tandem Bacteriophytochromes.
Structure, 23, 2015
|
|
8K6Y
 
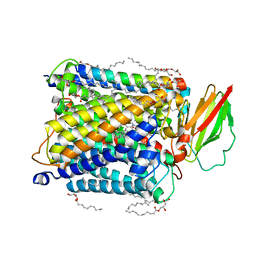 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
5DA5
 
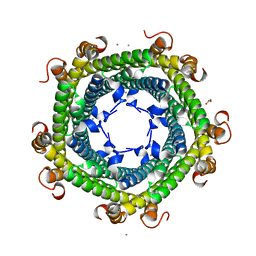 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 | | Descriptor: | CALCIUM ION, FE (III) ION, GLYCOLIC ACID, ... | | Authors: | He, D, Vanden Hehier, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
8K65
 
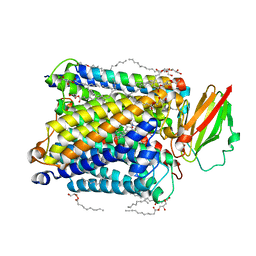 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
1YC7
 
 | | cAbAn33 VHH fragment against VSG | | Descriptor: | SULFATE ION, anti-VSG immunoglobulin heavy chain variable domain cAbAn33 | | Authors: | Conrath, K, Vincke, C, Stijlemans, B, Schymkowitz, J, Wyns, L, Muyldermans, S, Loris, R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Antigen Binding and Solubility Effects upon the Veneering of a Camel VHH in Framework-2 to Mimic a VH.
J.Mol.Biol., 350, 2005
|
|
3QMH
 
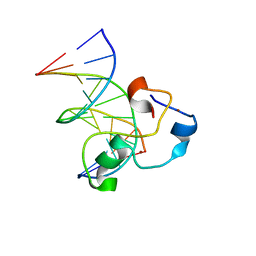 | | Structural Basis of Selective Binding of Non-Methylated CpG islands (DNA-TCGA) by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*TP*CP*GP*AP*TP*GP*GP*C)-3', CpG-binding protein, ZINC ION | | Authors: | Xu, C, Bian, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
1YGK
 
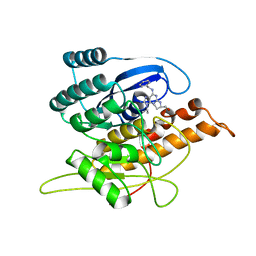 | | Crystal Structure of Pyridoxal Kinase in Complex with Roscovitine and Derivatives | | Descriptor: | Pyridoxal kinase, R-ROSCOVITINE | | Authors: | Tang, L, Li, M.-H, Cao, P, Wang, F, Chang, W.-R, Bach, S, Reinhardt, J, Ferandin, Y, Koken, M, Galons, H, Wan, Y, Gray, N, Meijer, L, Jiang, T, Liang, D.-C. | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives
J.Biol.Chem., 280, 2005
|
|
4QT6
 
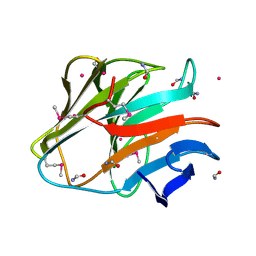 | | Crystal structure of the SPRY domain of human HERC1 | | Descriptor: | FORMAMIDE, Probable E3 ubiquitin-protein ligase HERC1, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Guan, X, Wernimont, A, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2015-01-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the SPRY domain of human HERC1
To be Published
|
|
4QTT
 
 | | Structure of S. cerevisiae Bud23-Trm112 complex involved in formation of m7G1575 on 18S rRNA (apo-form) | | Descriptor: | 1,2-ETHANEDIOL, DODECAETHYLENE GLYCOL, Multifunctional methyltransferase subunit TRM112, ... | | Authors: | Letoquart, J, Huvelle, E, Wacheul, L, Bourgeois, G, Zorbas, C, Graille, M, Heurgue-Hamard, V, Lafontaine, D.L.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-24 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
