2XVS
 
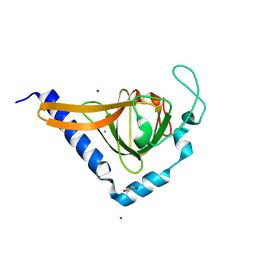 | | Crystal structure of human TTC5 (Strap) C-terminal OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Adams, J, Pike, A.C.W, Maniam, S, Sharpe, T.D, Coutts, A.S, Knapp, S, La Thangue, B, Bullock, A.N. | | Deposit date: | 2010-10-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XDV
 
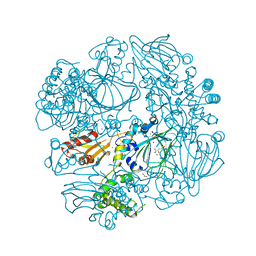 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | Authors: | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-07 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4PTI
 
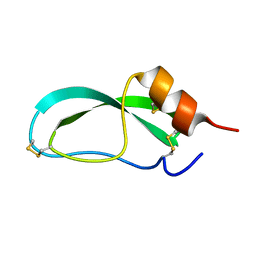 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
6LIL
 
 | |
7OQH
 
 | | CryoEM structure of the transcription termination factor Rho from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcription termination factor Rho | | Authors: | Saridakis, E, Vishwakarma, R, Lai Kee Him, J, Martin, K, Simon, I, Cohen-Gonsaud, M, Coste, F, Bron, P, Margeat, E, Boudvillain, M. | | Deposit date: | 2021-06-03 | | Release date: | 2022-02-09 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism.
Commun Biol, 5, 2022
|
|
4Q1E
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 10 {2-{[(1R/S)-1-{2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-methyl-1,3-thiazol 4-yl}ethyl]sulfanyl}pyrimidine-4,6-diamine} | | Descriptor: | (R)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-methylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, (S)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-methylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, Deoxycytidine kinase, ... | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
8OKA
 
 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
2Z1S
 
 | | Beta-glucosidase B from paenibacillus polymyxa complexed with cellotetraose | | Descriptor: | Beta-glucosidase B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Isorna, P, Sanz-Aparicio, J. | | Deposit date: | 2007-05-12 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
6I1A
 
 | | Crystal structure of rutinosidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, rutinosidase | | Authors: | Pachl, P, Rezacova, P, Kapesova, J. | | Deposit date: | 2018-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rutinosidase from Aspergillus niger: crystal structure and insight into the enzymatic activity.
Febs J., 287, 2020
|
|
6RVH
 
 | | NADH-dependent Coenzyme A Disulfide Reductase soaked with Menadione | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, ... | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6RUZ
 
 | | NADH-dependent Coenzyme A Disulfide Reductase | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
2Z3C
 
 | |
6IJL
 
 | |
6RVO
 
 | |
4CVN
 
 | | Structure of the Fap7-Rps14 complex | | Descriptor: | 30S RIBOSOMAL PROTEIN S11, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Loch, J, Blaud, M, Rety, S, Lebaron, S, Deschamps, P, Bareille, J, Jombart, J, Robert-Paganin, J, Delbos, L, Chardon, F, Zhang, E, Charenton, C, Tollervey, D, Leulliot, N. | | Deposit date: | 2014-03-28 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | RNA Mimicry by the Fap7 Adenylate Kinase in Ribosome Biogenesis
Plos Biol., 12, 2014
|
|
6LE5
 
 | | Crystal structure of the mitochondrial calcium uptake 1 and 2 heterodimer (MICU1-MICU2 heterodimer) in an apo state | | Descriptor: | Calcium uptake protein 1, mitochondrial, Calcium uptake protein 2 | | Authors: | Park, J, Lee, Y, Park, T, Kang, J.Y, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2019-11-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift.
Iucrj, 7, 2020
|
|
4ZNH
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Fluoro-substituted OBHS derivative | | Descriptor: | 2-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3HI6
 
 | |
4ZNT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Bromo-substituted OBHS derivative | | Descriptor: | 3-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZK4
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with 7-(5-isopropoxy-pyridin-3-yl)-1-methyl-1,7-diaza-spiro[4.4]nonane | | Descriptor: | (5R)-1-methyl-7-[5-(propan-2-yloxy)pyridin-3-yl]-1,7-diazaspiro[4.4]nonane, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bobango, J, Wu, J, Talley, T.T. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with 7-(5-isopropoxy-pyridin-3-yl)-1-methyl-1,7-diaza-spiro[4.4]nonane.
To be Published
|
|
4Q1A
 
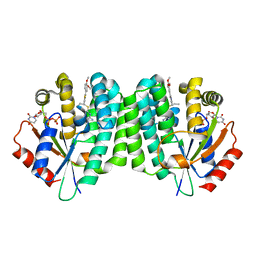 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 6 {2-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]ethanol} | | Descriptor: | 2-(5-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenoxy)ethan-1-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
2Z3B
 
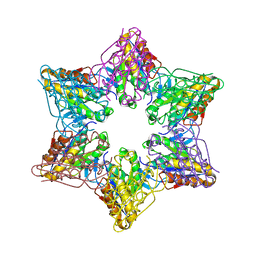 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
4GG8
 
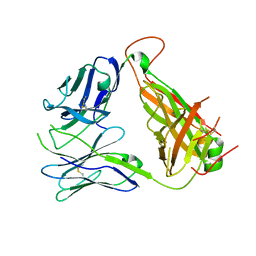 | | Immune Receptor | | Descriptor: | T-CELL RECEPTOR, SP3.4 ALPHA CHAIN, SP3.4 BETA CHAIN, ... | | Authors: | Broughton, S.E, Theodossis, A, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease.
Immunity, 37, 2012
|
|
2Z3D
 
 | |
