8A5B
 
 | | Crystal structure of human cathepsin L in complex with covalently bound MG-101 | | Descriptor: | Calpain Inhibitor I, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
5M1J
 
 | | Nonstop ribosomal complex bound with Dom34 and Hbs1 | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Hilal, T, Yamamoto, H, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution.
Nat Commun, 7, 2016
|
|
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZVF
 
 | | Crystal structure of human cathepsin L in complex with covalently bound CLIK148 | | Descriptor: | (2S)-N-[(2S)-1-(dimethylamino)-1-oxidanylidene-3-phenyl-propan-2-yl]-2-oxidanyl-N'-(2-pyridin-2-ylethyl)butanediamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Tsuge, H, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
2VAS
 
 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
8V8Z
 
 | |
6B97
 
 | | Crystal structure of PDE2 in complex with complex 9 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1-methyl-N-{(1R)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The identification of a novel lead class for phosphodiesterase 2 inhibition by fragment-based drug design.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8V9B
 
 | | Lipoprotein(a) Kringle IV domain 7 - Lp(a) KIV7 in complex with LY3441732 | | Descriptor: | (2S,2'S)-3,3'-[carbonylbis(azanediyl-3,1-phenylene)]bis{2-[(3R)-pyrrolidin-1-ium-3-yl]propanoate}, Apolipoprotein(a), MAGNESIUM ION, ... | | Authors: | Hendle, J, Weichert, K, Sauder, J.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Discovery of potent small-molecule inhibitors of lipoprotein(a) formation.
Nature, 629, 2024
|
|
5ITQ
 
 | | Crystal Structure of Human NEIL1, Free Protein | | Descriptor: | Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ITY
 
 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing Thymine Glycol | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1, GLYCEROL | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IS7
 
 | | Crystal structure of mouse CARM1 in complex with decarboxylated SAH | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(3-aminopropyl)-5'-thioadenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with decarboxylated SAH
To Be Published
|
|
5ISI
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine) | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 5'-amino-5'-deoxyadenosine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0920 (5'-amino-5'-deoxyadenosine)
To Be Published
|
|
6W8W
 
 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
5ISC
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0491 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0491
To Be Published
|
|
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
2VLD
 
 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
5NFZ
 
 | | TUBULIN-MTC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-(2,3,4-trimethoxyphenyl)cyclohepta-2,4,6-trien-1-one, CALCIUM ION, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
8X51
 
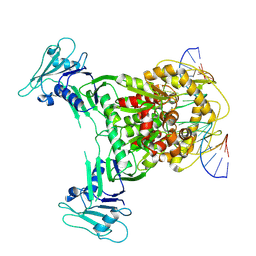 | | Cryo-EM structure of Gabija GajA in complex with DNA(focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6VW0
 
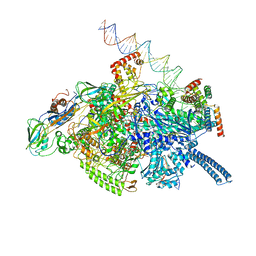 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
5AP9
 
 | | Controlled lid-opening in Thermomyces lanuginosus lipase - a switch for activity and binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Skjold-Joergensen, J, Vind, J, Moroz, O.V, Blagova, E.V, Bhatia, V.K, Svendsen, A, Wilson, K.S, Bjerrum, M.J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function.
Biochim. Biophys. Acta, 1865, 2017
|
|
5N9S
 
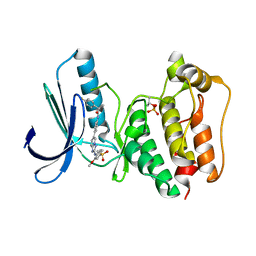 | | TTK kinase domain in complex with BAY 1161909 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, Dual specificity protein kinase TTK | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5JFX
 
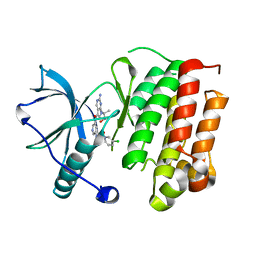 | | Crystal structure of TrkA in complex with PF-06273340 | | Descriptor: | High affinity nerve growth factor receptor, N-{5-[2-amino-7-(1-hydroxy-2-methylpropan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}-2-(5-chloropyridin-2-yl)acetamide | | Authors: | Jayasankar, J, Kurumbail, R, Skerratt, S, Brown, D. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
