4NN3
 
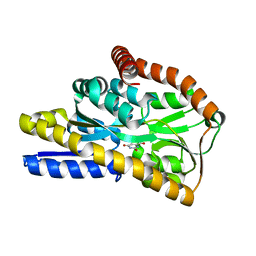 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens (Desal_2161), Target EFI-510109, with bound orotic acid | | Descriptor: | CHLORIDE ION, OROTIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-16 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3KIP
 
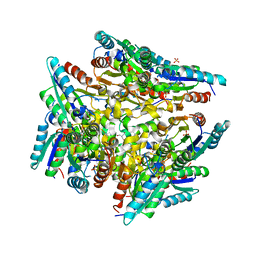 | | Crystal structure of type-II 3-dehydroquinase from C. albicans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinase, type II, ... | | Authors: | Trapani, S, Schoehn, G, Navaza, J, Abergel, C. | | Deposit date: | 2009-11-02 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KKZ
 
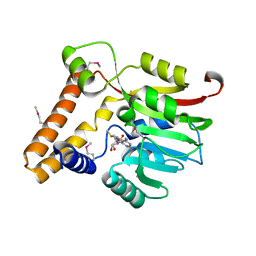 | | Crystal structure of the Q5LES9_BACFN protein from Bacteroides fragilis. Northeast Structural Genomics Consortium Target BfR250. | | Descriptor: | S-ADENOSYLMETHIONINE, uncharacterized protein Q5LES9 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Crystal structure of the Q5LES9_BACFN protein from Bacteroides fragilis.
To be Published
|
|
3WWJ
 
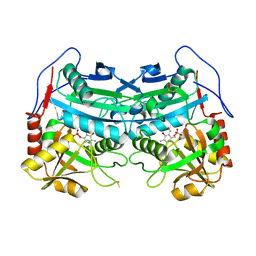 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3ZPZ
 
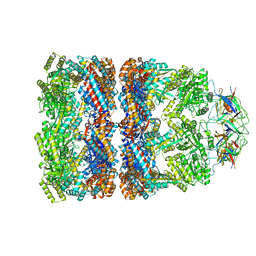 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
3KN3
 
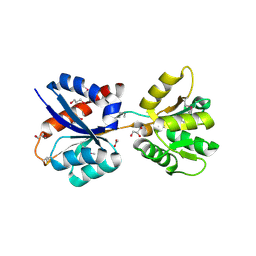 | | Crystal Structure of LysR Substrate Binding Domain (25-263) of Putative Periplasmic Protein from Wolinella succinogenes | | Descriptor: | ACETIC ACID, CITRIC ACID, GLUTATHIONE, ... | | Authors: | Kim, Y, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-01 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structure of LysR Substrate Binding Domain from Wolinella succinogenes
To be Published
|
|
4NQ8
 
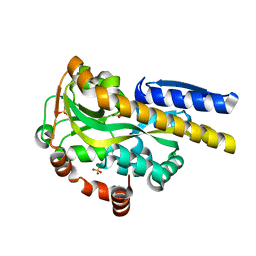 | | Crystal structure of a trap periplasmic solute binding protein from Bordetella bronchispeptica (bb3421), target EFI-510039, with density modeled as pantoate | | Descriptor: | CHLORIDE ION, PANTOATE, Putative periplasmic substrate-binding transport protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-24 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3KNB
 
 | |
3KOJ
 
 | | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp. Northeast Structural Genomics Consortium Target SnR59a. | | Descriptor: | uncharacterized protein ycf41 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Wang, H, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp.
To be Published
|
|
3KQ7
 
 | |
3X05
 
 | | Crystal structure of PIP4KIIBETA T201M complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X06
 
 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3KSL
 
 | | Structure of FPT bound to DATFP-DH-GPP | | Descriptor: | (2S,6E)-8-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}-2,6-dimethyloct-6-en-1-yl (2S)-3,3,3-trifluoro-2-hydrazinopropanoate, Farnesyltransferase, CAAX box, ... | | Authors: | Hovlid, M.L, Edelstein, R.L, Henry, O, Ochocki, J, DeGraw, A, Lenevich, S, Talbot, T, Young, V, Hruza, A.W, Lopez-Gallego, F, Labello, N.P, Strickland, C.L, Schmidt-Dannert, C, Distefano, M.D. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis, properties, and applications of diazotrifluropropanoyl-containing photoactive analogs of farnesyl diphosphate containing modified linkages for enhanced stability.
Chem.Biol.Drug Des., 75, 2010
|
|
3KT6
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with Trp | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
3X3B
 
 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
3KUG
 
 | |
3KUE
 
 | | Crystal structure of E. coli HPPK(E77A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-11-27 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
4B8A
 
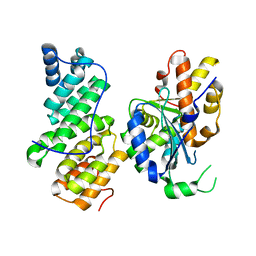 | | Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4O04
 
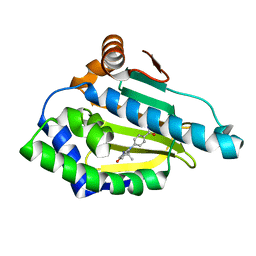 | | Identification of novel HSP90/isoform selective inhibitors using structure-based drug design. Demonstration of potential utility in treating CNS disorders such as Huntington's disease | | Descriptor: | 4-(2,7,7-trimethyl-5-oxo-1,2,3,4,5,6,7,8-octahydro-9H-beta-carbolin-9-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Zuccola, H.J, Ernst, J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of Novel HSP90 alpha / beta Isoform Selective Inhibitors Using Structure-Based Drug Design. Demonstration of Potential Utility in Treating CNS Disorders such as Huntington's Disease.
J.Med.Chem., 57, 2014
|
|
3KYE
 
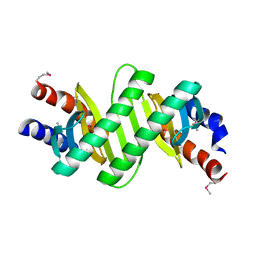 | | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis | | Descriptor: | Roadblock/LC7 domain, Robl_LC7 | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-05 | | Release date: | 2009-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis
To be Published
|
|
4B0S
 
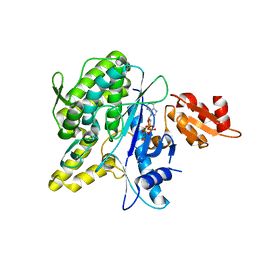 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4B6F
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1QGI
 
 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
4B7N
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
3L49
 
 | | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER SUBUNIT FROM Rhodobacter sphaeroides 2.4.1 | | Descriptor: | ABC sugar (Ribose) transporter, periplasmic substrate-binding subunit, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Do, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER FROM Rhodobacter sphaeroides
To be Published
|
|
