4JZV
 
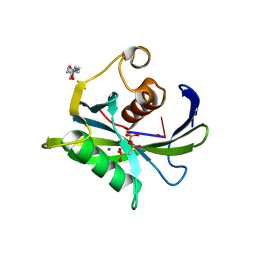 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | 著者 | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | 登録日 | 2013-04-03 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZJP
 
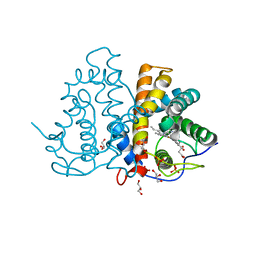 | | M.acetivorans protoglobin in complex with imidazole | | 分子名称: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | 著者 | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | 登録日 | 2014-06-02 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3ZYE
 
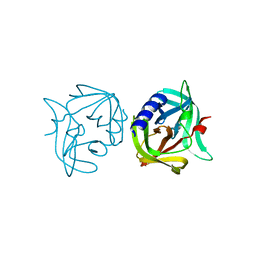 | | Crystal structure of 3C protease mutant (T68A) of coxsackievirus B3 | | 分子名称: | 3C PROTEINASE | | 著者 | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-08-22 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | 著者 | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | 登録日 | 2013-02-12 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3JAF
 
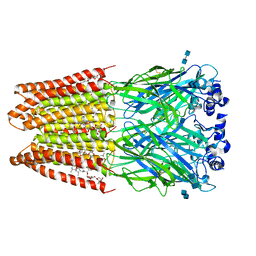 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine/ivermectin-bound state | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | 著者 | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | 登録日 | 2015-06-08 | | 公開日 | 2015-09-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.801 Å) | | 主引用文献 | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | 著者 | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | 登録日 | 2011-08-11 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZGX
 
 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | 分子名称: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | 著者 | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | 登録日 | 2012-12-19 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3ZU5
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZII
 
 | | Bacillus subtilis SepF G109K, C-terminal domain | | 分子名称: | CELL DIVISION PROTEIN SEPF | | 著者 | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | 登録日 | 2013-01-09 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZKE
 
 | | Structure of LC8 in complex with Nek9 peptide | | 分子名称: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | 著者 | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | 登録日 | 2013-01-22 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
3J9V
 
 | | Yeast V-ATPase state 3 | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | 登録日 | 2015-02-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.3 Å) | | 主引用文献 | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3JCZ
 
 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-27 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JA8
 
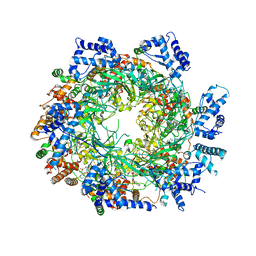 | | Cryo-EM structure of the MCM2-7 double hexamer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | 著者 | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | 登録日 | 2015-05-09 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
3JD3
 
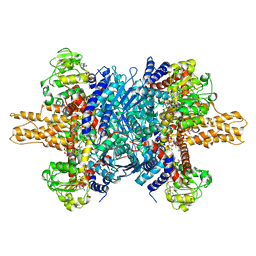 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3ZRV
 
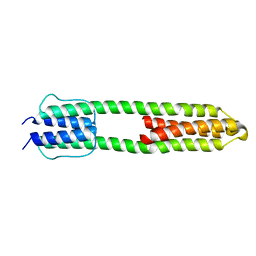 | | The high resolution structure of a dimeric Hamp-Dhp fusion displays asymmetry - A291F mutant | | 分子名称: | HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | | 著者 | Zeth, K, Hulko, M, Ferris, H.U, Martin, J. | | 登録日 | 2011-06-20 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
3JB9
 
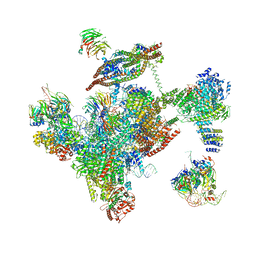 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | 登録日 | 2015-08-09 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3ZYQ
 
 | | Crystal structure of the tandem VHS and FYVE domains of Hepatocyte growth factor-regulated tyrosine kinase substrate (HGS-Hrs) at 1.48 A resolution | | 分子名称: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, SULFATE ION, ... | | 著者 | Muniz, J.R.C, Ayinampudi, V, Shrestha, L, Krojer, T, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | 登録日 | 2011-08-24 | | 公開日 | 2011-09-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) at 1.48 A Resolution
To be Published
|
|
1SW0
 
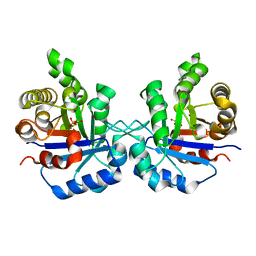 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | 著者 | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | 登録日 | 2004-03-30 | | 公開日 | 2004-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
3ZJL
 
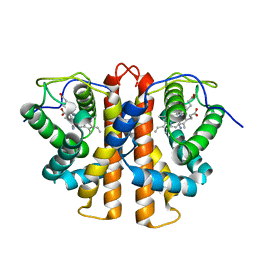 | | Leu(142)G4Ala mutation of M.acetivorans protoglobin in complex with cyanide | | 分子名称: | CYANIDE ION, PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
4L7Z
 
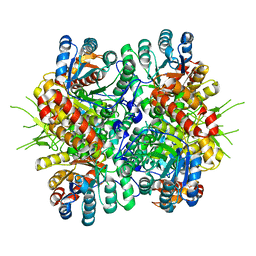 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | 著者 | Zarzycki, J, Kerfeld, C.A. | | 登録日 | 2013-06-14 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
3ZJQ
 
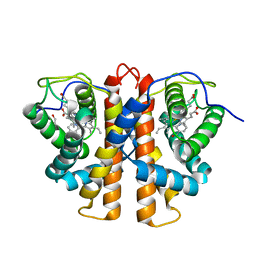 | | M.acetivorans protoglobin in complex with nicotinamide | | 分子名称: | GLYCEROL, NICOTINAMIDE, PROTOGLOBIN, ... | | 著者 | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
1SZJ
 
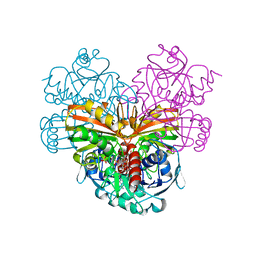 | |
3ZMV
 
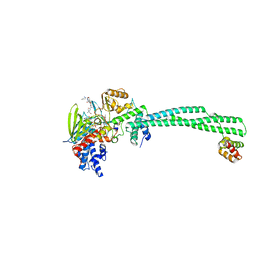 | | LSD1-CoREST in complex with PLSFLV peptide | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | 著者 | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | 登録日 | 2013-02-12 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
