3VSV
 
 | | The complex structure of XylC with xylose | | 分子名称: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | 著者 | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | 登録日 | 2012-05-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3IB6
 
 | | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b | | 分子名称: | Uncharacterized protein | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-15 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b
To be Published
|
|
3VY8
 
 | | Crystal Structure of PorB from Neisseria meningitidis in complex with Cesium ion, space group P63 | | 分子名称: | CESIUM ION, Outer membrane protein | | 著者 | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | 登録日 | 2012-09-21 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
3W08
 
 | | Crystal structure of aldoxime dehydratase | | 分子名称: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hashimoto, H, Nomura, J, Hashimoto, Y, Oinuma, K.I, Wada, K, Hishiki, A, Hara, K, Kobayashi, M. | | 登録日 | 2012-10-24 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3IGP
 
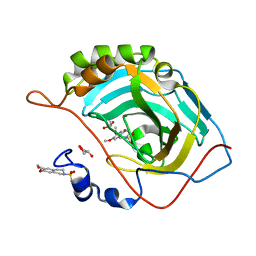 | | Structure of inhibitor binding to Carbonic Anhydrase II | | 分子名称: | 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Gitto, R, Agnello, S, Brynda, J, Mader, P, Supuran, C.T, Chimirri, A. | | 登録日 | 2009-07-28 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Identification of 3,4-Dihydroisoquinoline-2(1H)-sulfonamides as Potent Carbonic Anhydrase Inhibitors: Synthesis, Biological Evaluation, and Enzyme-Ligand X-ray Studies.
J.Med.Chem., 2010
|
|
3V7P
 
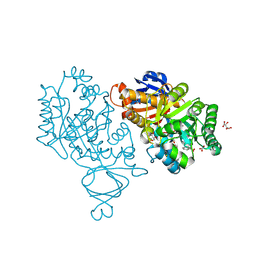 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | 分子名称: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-12-21 | | 公開日 | 2012-01-11 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
3IKJ
 
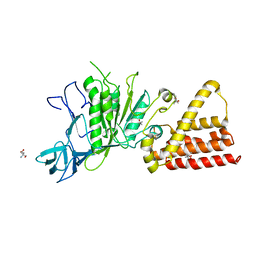 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | 著者 | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-08-06 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3V7L
 
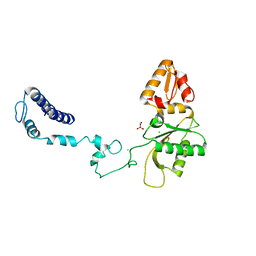 | | Apo Structure of Rat DNA polymerase beta K72E variant | | 分子名称: | CHLORIDE ION, DNA polymerase beta, SODIUM ION, ... | | 著者 | Rangarajan, S, Jaeger, J. | | 登録日 | 2011-12-21 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
4M0E
 
 | | Structure of human acetylcholinesterase in complex with dihydrotanshinone I | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cheung, J, Gary, E.N, Shiomi, K, Rosenberry, T.L. | | 登録日 | 2013-08-01 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of human acetylcholinesterase bound to dihydrotanshinone I and territrem B show peripheral site flexibility.
ACS Med Chem Lett, 4, 2013
|
|
1RZW
 
 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | 分子名称: | Protein AF2095(GR4) | | 著者 | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-12-29 | | 公開日 | 2004-11-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
3VBW
 
 | |
3ILL
 
 | |
3IPF
 
 | | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR8c. | | 分子名称: | uncharacterized protein | | 著者 | Vorobiev, S.M, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Andre, I, Rossi, P, Kennedy, M, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-08-17 | | 公開日 | 2009-09-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.988 Å) | | 主引用文献 | Crystal structure of the Q251Q8_DESHY protein from Desulfitobacterium hafniense.
To be Published
|
|
3IPO
 
 | | Crystal structure of YnjE | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | 著者 | Haenzelmann, P, Kuper, J, Schindelin, H. | | 登録日 | 2009-08-18 | | 公開日 | 2009-12-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3VBD
 
 | | Complex of human carbonic anhydrase II with 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide | | 分子名称: | 4-(6-methoxy-3,4-dihydroisoquinolin-1-yl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | 著者 | Mader, P, Brynda, J, Rezacova, P. | | 登録日 | 2012-01-02 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Synthesis, Structure-Activity Relationship Studies, and X-ray Crystallographic Analysis of Arylsulfonamides as Potent Carbonic Anhydrase Inhibitors.
J.Med.Chem., 55, 2012
|
|
3VI0
 
 | |
3IQZ
 
 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methylene-tetrahydromethanopterin | | 分子名称: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, CALCIUM ION, F420-dependent methylenetetrahydromethanopterin dehydrogenase, ... | | 著者 | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | 登録日 | 2009-08-21 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
3IFE
 
 | | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'. | | 分子名称: | Peptidase T, SODIUM ION, SULFATE ION, ... | | 著者 | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-07-24 | | 公開日 | 2009-08-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
1N8Q
 
 | | LIPOXYGENASE IN COMPLEX WITH PROTOCATECHUIC ACID | | 分子名称: | 3,4-DIHYDROXYBENZOIC ACID, FE (II) ION, lipoxygenase-3 | | 著者 | Borbulevych, O.Y, Jankun, J, Selman, S.H, Skrzypczak-Jankun, E. | | 登録日 | 2002-11-21 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Lipoxygenase interactions with natural flavonoid, quercetin, reveal a complex with protocatechuic acid in its X-ray structure at 2.1 A resolution.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3VPJ
 
 | |
1SH0
 
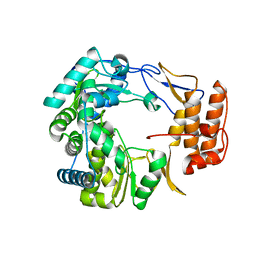 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | 分子名称: | RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
3VP3
 
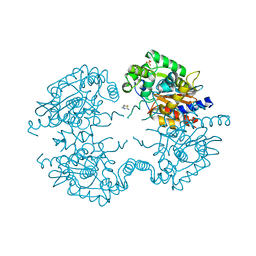 | | Crystal structure of human glutaminase in complex with inhibitor 3 | | 分子名称: | 5,5'-pentane-1,5-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | 著者 | Thangavelu, K, Sivaraman, J. | | 登録日 | 2012-02-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VP4
 
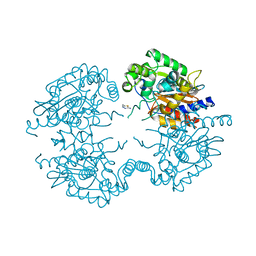 | | Crystal structure of human glutaminase in complex with inhibitor 4 | | 分子名称: | 5,5'-butane-1,4-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial | | 著者 | Thangavelu, K, Sivaraman, J. | | 登録日 | 2012-02-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IPP
 
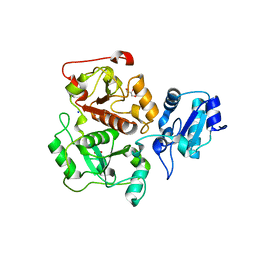 | | crystal structure of sulfur-free YnjE | | 分子名称: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | 著者 | Haenzelmann, P, Kuper, J, Schindelin, H. | | 登録日 | 2009-08-18 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | 分子名称: | Triosephosphate isomerase | | 著者 | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | 登録日 | 2004-03-18 | | 公開日 | 2004-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
