2A43
 
 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
2ABM
 
 | | Crystal Structure of Aquaporin Z Tetramer Reveals both Open and Closed Water-conducting Channels | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Jiang, J, Daniels, B.V, Fu, D. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of AqpZ Tetramer Reveals Two Distinct Arg-189 Conformations Associated with Water Permeation through the Narrowest Constriction of the Water-conducting Channel.
J.Biol.Chem., 281, 2006
|
|
2AAV
 
 | | Solution NMR structure of Filamin A domain 17 | | Descriptor: | Filamin A | | Authors: | Nakamura, F, Pudas, R, Heikkinen, O, Permi, P, Kilpelainen, I, Munday, A.D, Hartwig, J.H, Stossel, T.P, Ylanne, J. | | Deposit date: | 2005-07-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the GPIb-filamin A complex
Blood, 107, 2006
|
|
3TQC
 
 | | Structure of the pantothenate kinase (coaA) from Coxiella burnetii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Pantothenate kinase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQE
 
 | | Structure of the malonyl CoA-acyl carrier protein transacylase (fabD) from Coxiella burnetii | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, Malonyl-CoA-[acyl-carrier-protein] transacylase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQY
 
 | | Structure of a single-stranded DNA-binding protein (ssb), from Coxiella burnetii | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRF
 
 | | Structure of a shikimate kinase (aroK) from Coxiella burnetii | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3UCD
 
 | | Asymmetric complex of human neuron specific enolase-2-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-10-26 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
1HQU
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | Authors: | Hsiou, Y, Ding, J, Arnold, E. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
6HNN
 
 | | Crystal structure of wild-type IdmH, a putative polyketide cyclase from Streptomyces antibioticus | | Descriptor: | Putative polyketide cyclase IdmH | | Authors: | Drulyte, I, Obajdin, J, Trinh, C, Hemsworth, G.R, Berry, A. | | Deposit date: | 2018-09-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase.
Iucrj, 6, 2019
|
|
1CJ1
 
 | | GROWTH FACTOR RECEPTOR BINDING PROTEIN SH2 DOMAIN (HUMAN) COMPLEXED WITH A PHOSPHOTYROSYL DERIVATIVE | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2), [1-[1-(6-CARBAMOYL-CYCLOHEX-2-ENYLCARBAMOYL)-CYCLOHEXYLCARBAMOYL]-2-(4-PHOSPHONOOXY-PHENYL)- ETHYL]-CARBAMIC ACID 3-AMINOBENZYLESTER | | Authors: | Rahuel, J. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design, synthesis, and X-ray crystallography of a high-affinity antagonist of the Grb2-SH2 domain containing an asparagine mimetic.
J.Med.Chem., 42, 1999
|
|
5KBV
 
 | | Cryo-EM structure of GluA2 bound to antagonist ZK200775 at 6.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.G, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
1HT1
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
2QSW
 
 | | Crystal structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis | | Descriptor: | GLYCEROL, Methionine import ATP-binding protein metN 2, ZINC ION | | Authors: | Osipiuk, J, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structure of C-terminal domain of ABC transporter / ATP-binding protein from Enterococcus faecalis.
To be Published
|
|
5K2I
 
 | |
2QT7
 
 | | Crystallographic structure of the mature ectodomain of the human receptor-type protein-tyrosine phosphatase IA-2 at 1.30 Angstroms | | Descriptor: | CALCIUM ION, Receptor-type tyrosine-protein phosphatase-like N | | Authors: | Primo, M.E, Klinke, S, Sica, M.P, Goldbaum, F.A, Jakoncic, J, Poskus, E, Ermacora, M.R. | | Deposit date: | 2007-08-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the Mature Ectodomain of the Human Receptor-type Protein-tyrosine Phosphatase IA-2
J.Biol.Chem., 283, 2008
|
|
1CQ8
 
 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C6-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-HEXANOIC ACID, ASPARTATE AMINOTRANSFERASE (2.6.1.1) | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
6HXX
 
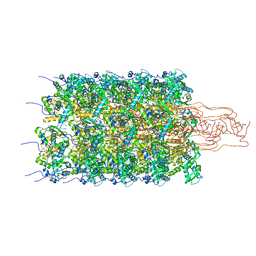 | | Potato virus Y | | Descriptor: | Coat protein, RNA (5'-R(P*UP*UP*UP*UP*U)-3') | | Authors: | Podobnik, M, Kezar, A, Novacek, J, Polak, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the multitasking nature of the potato virus Y coat protein.
Sci Adv, 5, 2019
|
|
2QUY
 
 | |
5OPT
 
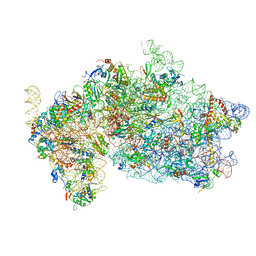 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
2QTO
 
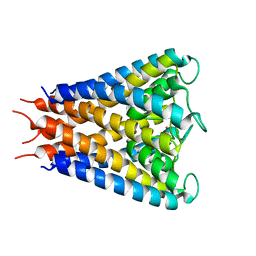 | | An anisotropic model for potassium channel KcsA | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Chen, X, Poon, B.K, Dousis, A, Wang, Q, Ma, J. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution
Structure, 15, 2007
|
|
2QV8
 
 | |
2QAI
 
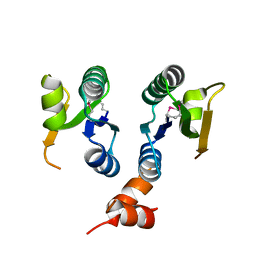 | | Crystal structure of the V-type ATP synthase subunit F from Pyrococcus furiosus. NESG target PfR7. | | Descriptor: | V-type ATP synthase subunit F | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Ma, L.-C, Shih, L, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-15 | | Release date: | 2007-06-26 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the V-type ATP synthase subunit F from Pyrococcus furiosus.
To be Published
|
|
5JVW
 
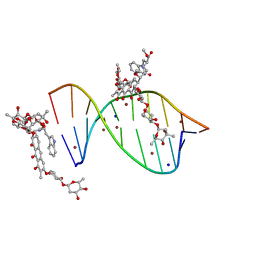 | | Crystal structure of mithramycin analogue MTM SA-Trp in complex with a 10-mer DNA AGAGGCCTCT. | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*GP*CP*CP*TP*CP*T)-3'), Plicamycin, mithramycin analogue MTM SA-Trp, ... | | Authors: | Hou, C, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2016-05-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1.
Nucleic Acids Res., 44, 2016
|
|
