1CN1
 
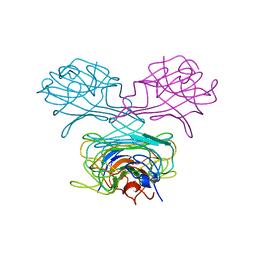 | | CRYSTAL STRUCTURE OF DEMETALLIZED CONCANAVALIN A. THE METAL-BINDING REGION | | Descriptor: | CONCANAVALIN A | | Authors: | Shoham, M, Yonath, A, Sussman, J.L, Moult, J, Traub, W, Gilboa(Kalb), A.J. | | Deposit date: | 1981-12-21 | | Release date: | 1982-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of demetallized concanavalin A: the metal-binding region.
J.Mol.Biol., 131, 1979
|
|
1CPT
 
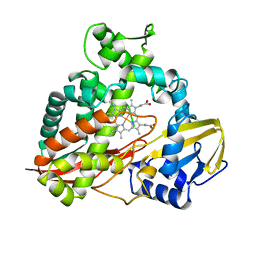 | | CRYSTAL STRUCTURE AND REFINEMENT OF CYTOCHROME P450-TERP AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME P450-TERP, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasemann, C.A, Ravichandran, K.G, Peterson, J.A, Deisenhofer, J. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and refinement of cytochrome P450terp at 2.3 A resolution.
J.Mol.Biol., 236, 1994
|
|
2AU1
 
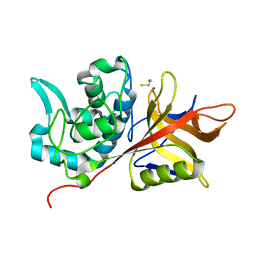 | | Crystal Structure of group A Streptococcus MAC-1 orthorhombic form | | Descriptor: | BETA-MERCAPTOETHANOL, IgG-degrading protease | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-26 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2AVO
 
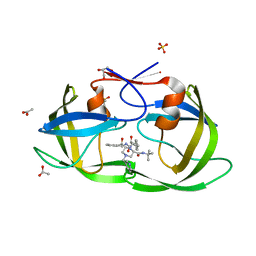 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AX6
 
 | | Crystal Structure Of The Androgen Receptor Ligand Binding Domain T877A Mutant In Complex With Hydroxyflutamide | | Descriptor: | Androgen receptor, HYDROXYFLUTAMIDE | | Authors: | Bohl, C.E, Miller, D.D, Chen, J, Bell, C.E, Dalton, J.T. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Accommodation of Nonsteroidal Ligands in the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
1JC7
 
 | |
4IXD
 
 | | X-ray structure of lfa-1 i-domain in complex with ibe-667 at 1.8a resolution | | Descriptor: | 4-(3-{4-[(3-aminopropyl)carbamoyl]phenyl}-1H-indazol-1-yl)-N-methylbenzamide, Integrin alpha-L, MAGNESIUM ION | | Authors: | Kallen, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and x-ray structure based investigation of an ICAM-1 binding enhancing small molecule activator of LFA-1
To be Published, 2013
|
|
2BCO
 
 | | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14 | | Descriptor: | Succinylglutamate desuccinylase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Benach, J, Zhou, W, Acton, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-19 | | Release date: | 2005-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14
To be Published
|
|
2B94
 
 | |
1OBV
 
 | | Y94F flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Romero, A, Ramon, A, Fernandez-Cabrera, C, Irun, M.P, Sancho, J. | | Deposit date: | 2003-01-31 | | Release date: | 2003-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Fmn Binds to Anabaena Apoflavodoxin: A Hydrophobic Encounter at an Open Binding Site
J.Biol.Chem., 278, 2003
|
|
2ZLG
 
 | | The Structual Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase | | Descriptor: | (5R,9S,12S,15S,18S,21S)-21-benzyl-12,18-bis(carboxymethyl)-15-cyclohexyl-1-(9H-fluoren-9-yl)-4-methyl-9-(2-methylpropyl)-3,6,10,13,16,19-hexaoxo-5-phenyl-2-oxa-4,8,11,14,17,20-hexaazadocosan-22-oic acid, GLYCEROL, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Fairman, J.W, Wijerathna, S.R, LaMacchia, J, Kreischer, N.R, Helmbrecht, E, Cooperman, B.S, Dealwis, C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Structural Basis for Peptidomimetic Inhibition of Eukaryotic Ribonucleotide Reductase: A Conformationally Flexible Pharmacophore
J.Med.Chem., 51, 2008
|
|
2ZM5
 
 | | Crystal structure of tRNA modification enzyme MiaA in the complex with tRNA(Phe) | | Descriptor: | MAGNESIUM ION, tRNA delta(2)-isopentenylpyrophosphate transferase, tRNA(Phe) | | Authors: | Sakai, J, Yao, M, Chimnaronk, S, Tanaka, I. | | Deposit date: | 2008-04-11 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon
Biochemistry, 48, 2009
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
5T5O
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, Lectin, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
6NDB
 
 | |
2ZQS
 
 | |
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | Authors: | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
6NRA
 
 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
2F91
 
 | | 1.2A resolution structure of a crayfish trypsin complexed with a peptide inhibitor, SGTI | | Descriptor: | CADMIUM ION, CHLORIDE ION, Serine protease inhibitor I/II, ... | | Authors: | Fodor, K, Harmat, V, Hetenyi, C, Kardos, J, Antal, J, Perczel, A, Patthy, A, Katona, G, Graf, L. | | Deposit date: | 2005-12-05 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme:Substrate Hydrogen Bond Shortening during the Acylation Phase of Serine Protease Catalysis.
Biochemistry, 45, 2006
|
|
2ZQV
 
 | |
2B89
 
 | |
2B88
 
 | |
2Y5D
 
 | | Crystal structure of C296A mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-12 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
2XBN
 
 | | Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MAGNESIUM ION, SERINE PALMITOYLTRANSFERASE | | Authors: | Lowther, J, Yard, B.A, Johnson, K.A, Carter, L.G, Bhat, V.T, Raman, M.C.C, Clarke, D.J, Ramakers, B, McMahon, S.A, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2010-04-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of the Plp-Dependent Enzyme Serine Palmitoyltransferase by Cycloserine: Evidence for a Novel Decarboxylative Mechanism of Inactivation.
Mol.Biosystems, 6, 2010
|
|
6NIY
 
 | | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | dal Maso, E, Glukhova, A, Zhu, Y, Garcia-Nafria, J, Tate, C.G, Atanasio, S, Reynolds, C.A, Ramirez-Aportela, E, Carazo, J.-M, Hick, C.A, Furness, S.G.B, Hay, D.L, Liang, Y.-L, Miller, L.J, Christopoulos, A, Wang, M.-W, Wootten, D, Sexton, P.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-01-23 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | The Molecular Control of Calcitonin Receptor Signaling.
Acs Pharmacol Transl Sci, 2, 2019
|
|
