3UGU
 
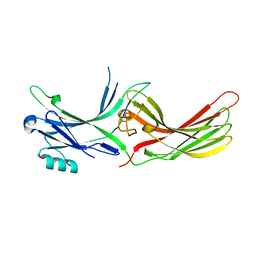 | |
6XJH
 
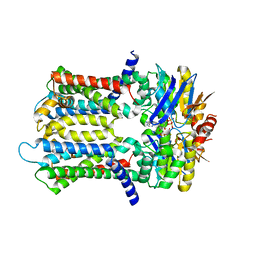 | | PmtCD ABC exporter without the basket domain at C2 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
1W8W
 
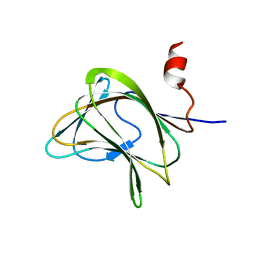 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1VYK
 
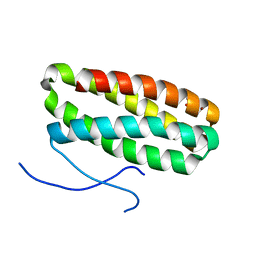 | | CRYSTAL STRUCTURE OF PSBQ PROTEIN OF PHOTOSYSTEM II FROM HIGHER PLANTS | | Descriptor: | ACETATE ION, OXYGEN-EVOLVING ENHANCER PROTEIN 3, ZINC ION | | Authors: | Hermoso, J.A, Balsera, M, De Las Rivas, J, Arellano, J.B. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.49 A Resolution Crystal Structure of Psbq from Photosystem II of Spinacia Oleracea Reveals a Ppii Structure in the N-Terminal Region.
J.Mol.Biol., 350, 2005
|
|
6XLT
 
 | |
1W59
 
 | | FtsZ dimer, empty (M. jannaschii) | | Descriptor: | CELL DIVISION PROTEIN FTSZ HOMOLOG 1, SULFATE ION | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1W4Z
 
 | | Structure of actinorhodin polyketide (actIII) Reductase | | Descriptor: | FORMIC ACID, KETOACYL REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Limpkin, C, Teartasin, W, Simpson, T.J, Crosby, J, Crump, M.P. | | Deposit date: | 2004-08-03 | | Release date: | 2004-12-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the Actiii Actinorhodin Polyketide Reductase; Proposed Mechanism for Acp and Polyketide Binding
Structure, 12, 2004
|
|
6XAG
 
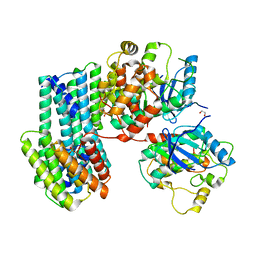 | | Apo BRAF dimer bound to 14-3-3 | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dimerization Induced by C-Terminal 14-3-3 Binding Is Sufficient for BRAF Kinase Activation.
Biochemistry, 59, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
1W5F
 
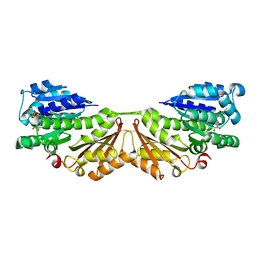 | | FtsZ, T7 mutated, domain swapped (T. maritima) | | Descriptor: | CELL DIVISION PROTEIN FTSZ, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
6XE0
 
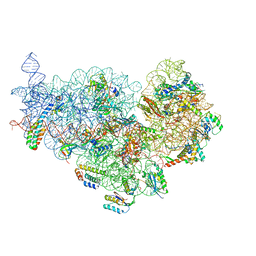 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
1W6F
 
 | | Arylamine N-acetyltransferase from Mycobacterium smegmatis with the anti-tubercular drug isoniazid bound in the active site. | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Holton, S, Fullham, E, Sim, E, Noble, M.E.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Anti-Tubercular Drug Isoniazid to the Arylamine N-Acetyltransferase Protein from Mycobacterium Smegmatis
Protein Sci., 14, 2005
|
|
3UM5
 
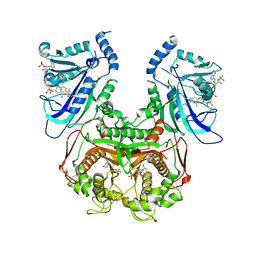 | | Double mutant (A16V+S108T) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-T9/94) complexed with pyrimethamine, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Kamchonwongpaisan, S, Chitnumsub, P, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
1W7A
 
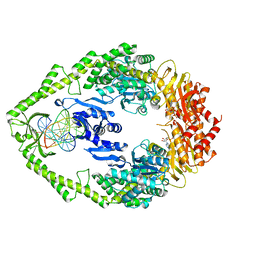 | | ATP bound MutS | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP* AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP* CP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lamers, M.H, Georgijevic, D, Lebbink, J, Winterwerp, H.H.K, Agianian, B, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
J.Biol.Chem., 279, 2004
|
|
1W90
 
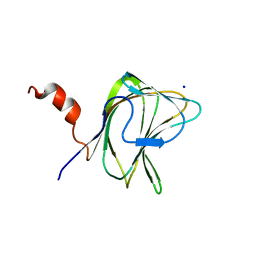 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
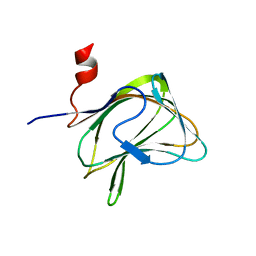 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
3UEM
 
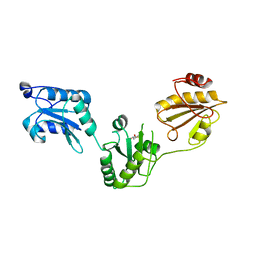 | | Crystal structure of human PDI bb'a' domains | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Protein disulfide-isomerase | | Authors: | Yu, J, Wang, C, Huo, L, Feng, W, Wang, C.-C. | | Deposit date: | 2011-10-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a'
J.Biol.Chem., 287, 2012
|
|
3UNL
 
 | | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2011-11-15 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni
To be Published
|
|
3UF2
 
 | |
6X60
 
 | | ClpP2 from Chlamydia trachomatis with resolved handle loop | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Azadmanesh, J, Struble, L.R, Seleem, M.A, Ouellette, S, Conda-Sheridan, M, Borgstahl, G.E.O. | | Deposit date: | 2020-05-27 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | ClpP2 from Chlamydia trachomatis with resolved handle loop
To Be Published
|
|
1W8B
 
 | | Factor7 - 413 complex | | Descriptor: | (S)-[(R)-2-(4-BENZYLOXY-3-METHOXY-PHENYL)-2-(4-CARBAMIMIDOYL-PHENYLAMINO)-ACETYLAMINO]-PHENYL-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION | | Authors: | Ackermann, J, Alig, L, Banner, D.W, Boehm, H.-J, Groebke-Zbinden, K, Hilpert, K, Lave, T, Kuehne, H, Obst-Sander, U, Riederer, M.A, Stahl, M, Tschopp, T.B, Weber, L, Wessel, H.P. | | Deposit date: | 2004-09-17 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective and Orally Bioavailable Phenylglycine Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1W28
 
 | | Conformational flexibility of the C-terminus with implications for substrate binding and catalysis in a new crystal form of deacetoxycephalosporin C synthase | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
