5AFH
 
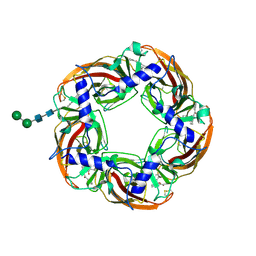 | | alpha7-AChBP in complex with lobeline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6GRY
 
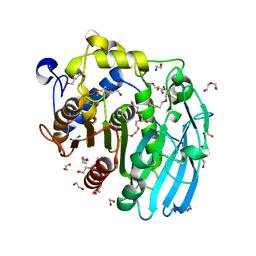 | | Glucuronoyl Esterase from Solibacter usitatus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | Authors: | Lo Leggio, L, Larsbrink, J, Meland Knudsen, R, Mazurkewich, S, Navarro Poulsen, J.C. | | Deposit date: | 2018-06-12 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.0003376 Å) | | Cite: | Biochemical and structural features of diverse bacterial glucuronoyl esterases facilitating recalcitrant biomass conversion.
Biotechnol Biofuels, 11, 2018
|
|
6GSG
 
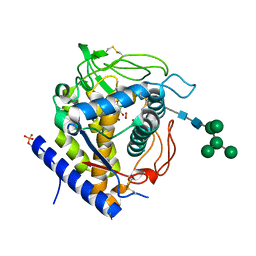 | | Crystal structure of Aspergillus oryzae catechol oxidase complexed with resorcinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Penttinen, L, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Unraveling Substrate Specificity and Catalytic Promiscuity of Aspergillus oryzae Catechol Oxidase.
Chembiochem, 19, 2018
|
|
2V5P
 
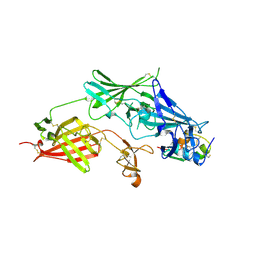 | | COMPLEX STRUCTURE OF HUMAN IGF2R DOMAINS 11-13 BOUND TO IGF-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, INSULIN-LIKE GROWTH FACTOR II, ... | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
6AWW
 
 | | Structure of PR 10 Allergen Ara h 8.01 in complex with 3-Hydroxy-2-naphthoic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Yarbrough, J, McBride, J, Hurlburt, B.K, Maleki, S.J, Pote, S.S, Chruszcz, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of PR-10 Allergen Ara h 8.01.
To Be Published
|
|
2VHR
 
 | |
3IAR
 
 | | The crystal structure of human adenosine deaminase | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, Adenosine deaminase, GLYCEROL, ... | | Authors: | Ugochukwu, E, Zhang, Y, Hapka, E, Yue, W.W, Bray, J.E, Muniz, J, Burgess-Brown, N, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The crystal structure of human adenosine deaminase
To be Published
|
|
4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
7PIM
 
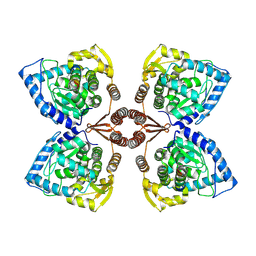 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6AUO
 
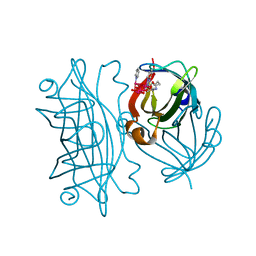 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112F | | Descriptor: | N-biotin-C-Co4(mu3-O)4(Py)3(H2O)4-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
6H0E
 
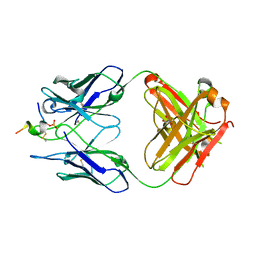 | | FAB dmCBTAU-22.1 IN COMPLEX WITH TAU PEPTIDE V1088-23 | | Descriptor: | GLYCEROL, HUMAN FAB ANTIBODY FRAGMENT OF dmCBTAU-22.1, Microtubule-associated protein tau | | Authors: | Juraszek, J, Steinbacher, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enhancement of therapeutic potential of a naturally occurring human antibody targeting a phosphorylated Ser422containing epitope on pathological tau.
Acta Neuropathol Commun, 6, 2018
|
|
5AFM
 
 | | alpha7-AChBP in complex with lobeline and fragment 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5-dibromo-N-(3-hydroxypropyl)-1H-pyrrole-2-carboxamide, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QS7
 
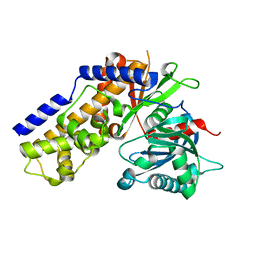 | |
6UOT
 
 | |
2JC5
 
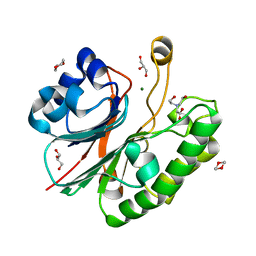 | | Apurinic Apyrimidinic (AP) endonuclease (NApe) from Neisseria Meningitidis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BICINE, EXODEOXYRIBONUCLEASE, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G.S, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
3FEO
 
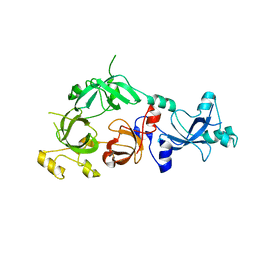 | | The crystal structure of MBTD1 | | Descriptor: | MBT domain-containing protein 1 | | Authors: | Amaya, M.F, Eryilmaz, J, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of a four-MBT repeat protein MBTD1.
Plos One, 4, 2009
|
|
6GJN
 
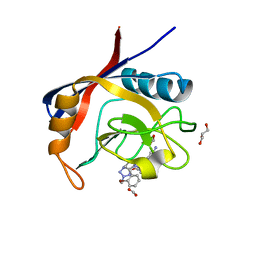 | | Cyclophilin A complexed with tri-vector ligand 15. | | Descriptor: | 1-[(4-aminophenyl)methyl]-3-[2-[(2~{R})-2-(2-bromophenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-1-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]urea, FORMIC ACID, GLYCEROL, ... | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
2JC4
 
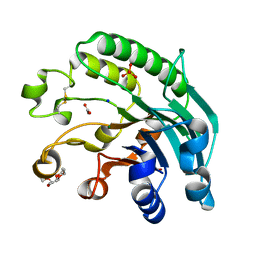 | | 3'-5' exonuclease (NExo) from Neisseria Meningitidis | | Descriptor: | ACETATE ION, DIHYDROGENPHOSPHATE ION, EXODEOXYRIBONUCLEASE III, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
5AFJ
 
 | | alpha7-AChBP in complex with lobeline and fragment 1 | | Descriptor: | (3S)-6-(4-bromophenyl)-3-hydroxy-1,3-dimethyl-2,3-dihydropyridin-4(1H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, ... | | Authors: | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6J31
 
 | | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin | | Descriptor: | (2E,2'E)-3,3'-(1,2-phenylene)di(prop-2-enoic acid), DBB-DSG-VAL-MEA-VAL-GLY-GLY-DVA-DLE, kcn28 | | Authors: | Shi, J, Liu, C.L, Zhang, B, Guo, W.J, Zhu, J.P, Xu, X, Xu, Q, Jiao, R.H, Tan, R.X, Ge, H.M. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Genome mining and biosynthesis of kitacinnamycins as a STING activator.
Chem Sci, 10, 2019
|
|
6AUE
 
 | | Artificial Metalloproteins Containing a Co4O4 Active Site - 2xm-S112Y-b | | Descriptor: | N-biotin-C-Co4(mu3-O)4(OAc)(Py)4(H2O)3-beta-alanine, Streptavidin | | Authors: | Olshansky, L, Vallapurakal, J, Huerta-Lavorie, R, Nguyen, A.I, Tilley, T.D, Borovik, A.S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Artificial Metalloproteins Containing Co
J. Am. Chem. Soc., 140, 2018
|
|
2VCI
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
6J5D
 
 | | Complex structure of MAb 4.2-scFv with louping ill virus envelope protein Domain III | | Descriptor: | Envelope, antibody heavy chain, antibody light chain | | Authors: | Yang, X, Qi, J, Peng, R, Dai, L, Gould, E.A, Tien, P, Gao, G.F. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of a Protective/Neutralizing Monoclonal Antibody Targeting Envelope Proteins of both Tick-Borne Encephalitis Virus and Louping Ill Virus.
J. Virol., 93, 2019
|
|
2VCJ
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-(5-chloro-2,4-dihydroxyphenyl)-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]isoxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
6IZ7
 
 | |
