5MZH
 
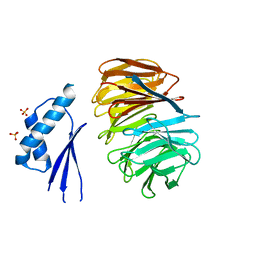 | | Crystal structure of ODA16 from Chlamydomonas reinhardtii | | Descriptor: | Dynein assembly factor with WDR repeat domains 1, SULFATE ION | | Authors: | Lorentzen, E, Taschner, M, Basquin, J, Mourao, A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46.
J. Biol. Chem., 292, 2017
|
|
5NAI
 
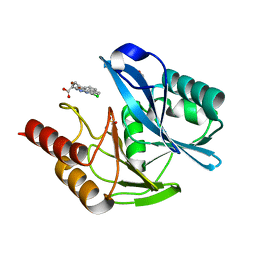 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-D-tryptophan (Compound 1) | | Descriptor: | (2~{R})-2-[(1-chloranyl-4-oxidanyl-isoquinolin-3-yl)carbonylamino]-3-(1~{H}-indol-3-yl)propanoic acid, Class B metallo-beta-lactamase, FLUORIDE ION, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
2W6G
 
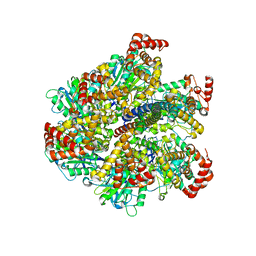 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 3. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Improving Diffraction by Humidity Control: A Novel Device Compatible with X-Ray Beamlines.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5AR8
 
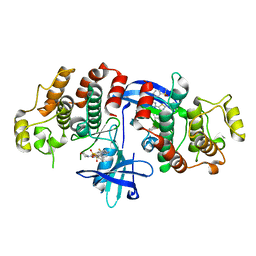 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biphenylsulfonamide | | Descriptor: | 2,6-bis(fluoranyl)-N-[3-[5-[2-[(3-methylsulfonylphenyl)amino]pyrimidin-4-yl]-2-morpholin-4-yl-1,3-thiazol-4-yl]phenyl]benzenesulfonamide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
4A8C
 
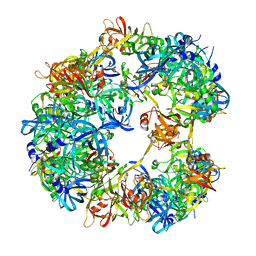 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
5BNO
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SLN | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
5N2Z
 
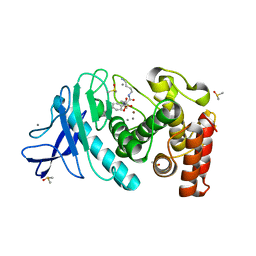 | | Thermolysin in complex with inhibitor JC286 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Thermolysin, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
5H0I
 
 | |
5H0S
 
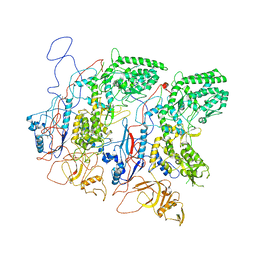 | | EM Structure of VP1A and VP1B | | Descriptor: | VP1 | | Authors: | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
5N3V
 
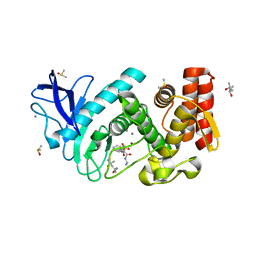 | | Thermolysin in complex with inhibitor JC292 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N70
 
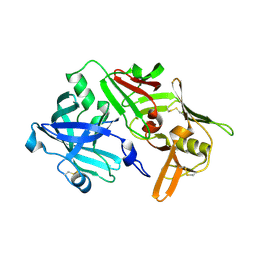 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) IN COMPLEX WITH THE N-TERMINAL OCTAPEPTIDE OF THE PROPEPTID | | Descriptor: | ALA-PHE-ARG-ILE-PRO-LEU-THR-ARG, Putative cathepsin d | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-17 | | Release date: | 2017-12-27 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
3JZR
 
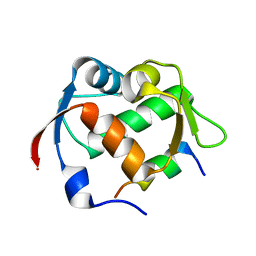 | |
4PQ8
 
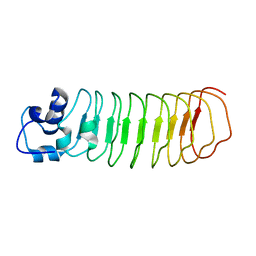 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
2WP8
 
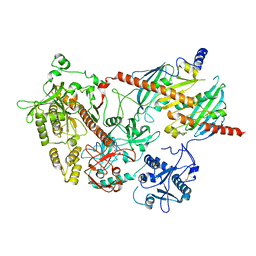 | | yeast rrp44 nuclease | | Descriptor: | CHLORIDE ION, EXOSOME COMPLEX COMPONENT RRP45, EXOSOME COMPLEX COMPONENT SKI6, ... | | Authors: | Basquin, J, Bonneau, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2009-08-03 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Yeast Exosome Functions as a Macromolecular Cage to Channel RNA Substrates for Degradation.
Cell(Cambridge,Mass.), 139, 2009
|
|
4PSJ
 
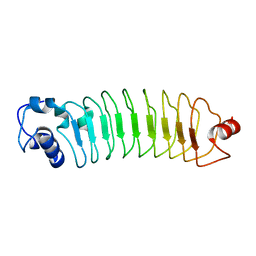 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR464. | | Descriptor: | OR464 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystal Structure of Engineered Protein OR464.
To be Published
|
|
5N55
 
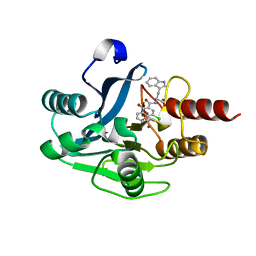 | | mono-Zinc VIM-5 metallo-beta-lactamase in complex with (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan (Compound 2) | | Descriptor: | (1-chloro-4-hydroxyisoquinoline-3-carbonyl)-L-tryptophan, Class B metallo-beta-lactamase, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
5H4Z
 
 | | Crystal structure of S202G mutant of human SYT-5 C2A domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Synaptotagmin-5 | | Authors: | Qiu, X, Ge, J, Yan, X, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2016-11-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural analysis of Ca(2+)-binding pocket of synaptotagmin 5 C2A domain
Int. J. Biol. Macromol., 95, 2017
|
|
4PRP
 
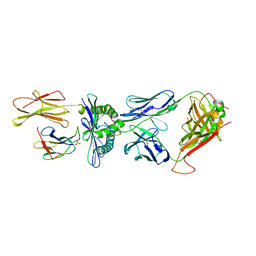 | | Crystal structure of TK3 TCR-HLA-B*35:01-HPVG-Q5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
5AML
 
 | |
5N7N
 
 | | CRYSTAL STRUCTURE OF CATHEPSIN D ZYMOGEN FROM THE TICK IXODES RICINUS (IRCD1) | | Descriptor: | AMMONIUM ION, Putative cathepsin d, SULFATE ION | | Authors: | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-27 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
4PTS
 
 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | Descriptor: | glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
5N4T
 
 | | VIM-2 metallo-beta-lactamase in complex with ((S)-3-mercapto-2-methylpropanoyl)-L-tryptophan (Compound 4) | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-[[(2~{S})-2-methyl-3-sulfanyl-propanoyl]amino]propanoic acid, BENZOIC ACID, Beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic analyses of isoquinoline complexes reveal a new mode of metallo-beta-lactamase inhibition.
Chem. Commun. (Camb.), 53, 2017
|
|
4AGT
 
 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with Fuc1-6GlcNAc. | | Descriptor: | FUCOSE-SPECIFIC LECTIN FLEA, SODIUM ION, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4AHA
 
 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with fucosylated monosaccharides (Fuc1-2Gal, Fuc1- 3GlcNAc, Fuc1-4GlcNAc and Fuc1-6GlcNAc) | | Descriptor: | Fucose-specific lectin, alpha-L-fucopyranose, alpha-L-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3JCB
 
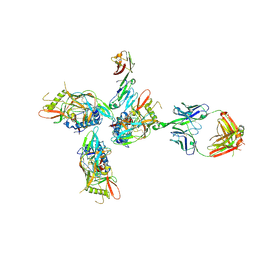 | | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | | Descriptor: | Antibody 36D5 heavy chain, Antibody 36D5 light chain, Envelope glycoprotein gp120, ... | | Authors: | Hu, G, Liu, J, Roux, K, Taylor, K.A. | | Deposit date: | 2015-11-25 | | Release date: | 2017-05-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5.
J. Virol., 91, 2017
|
|
