3BYI
 
 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | Descriptor: | Rho GTPase activating protein 15 | | Authors: | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
4OW3
 
 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
8JVD
 
 | |
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8JUC
 
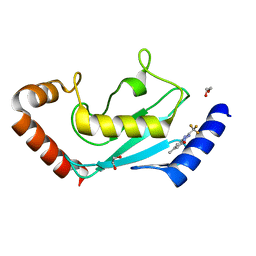 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
8K2O
 
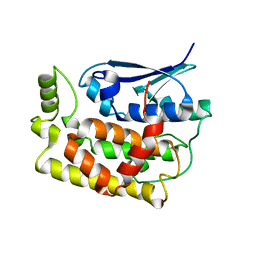 | | Crystal structure of Fhb7-M10 | | Descriptor: | Fhb7-M10 | | Authors: | Yang, J, Lei, X.G, Xiao, J.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2P
 
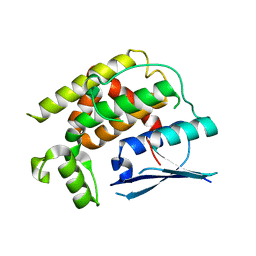 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
5N87
 
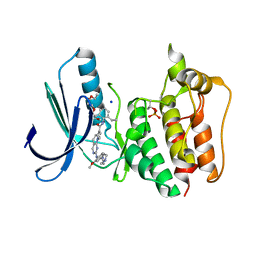 | | TTK kinase domain in complex with NTRC 0066-0 | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, Dual specificity protein kinase TTK, SODIUM ION, ... | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, Sterrenburg, J.G, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
6QYP
 
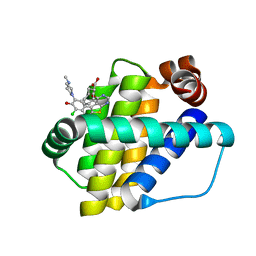 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
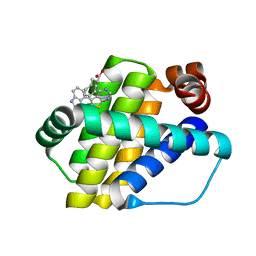 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYK
 
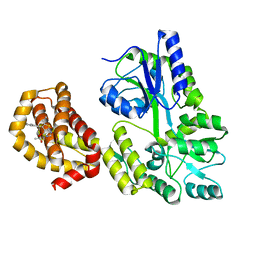 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6XKI
 
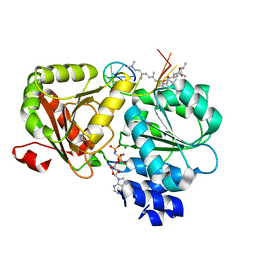 | | Crystal structure of eIF4A-I in complex with RNA bound to des-MePateA, a pateamine A analog | | Descriptor: | (3S,6Z,8E,11S,15R)-15-amino-3-[(1E,3E,5E)-7-(dimethylamino)-2,5-dimethylhepta-1,3,5-trien-1-yl]-9,11-dimethyl-4,12-dioxa-20-thia-21-azabicyclo[16.2.1]henicosa-1(21),6,8,18-tetraene-5,13-dione, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Liang, J, Naineni, S.K, Pelletier, J, Nagar, B. | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A.
Cell Chem Biol, 28, 2021
|
|
6QZ6
 
 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYN
 
 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
5XWR
 
 | | Crystal Structure of RBBP4-peptide complex | | Descriptor: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | Authors: | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GZ4
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-2 (TI-POST-2) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | Descriptor: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
6GZ5
 
 | | tRNA translocation by the eukaryotic 80S ribosome and the impact of GTP hydrolysis, Translocation-intermediate-POST-3 (TI-POST-3) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Flis, J, Holm, M, Rundlet, E.J, Loerke, J, Hilal, T, Dabrowski, M, Buerger, J, Mielke, T, Blanchard, S.C, Spahn, C.M.T, Budkevich, T.V. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis.
Cell Rep, 25, 2018
|
|
6WNC
 
 | |
2Y3G
 
 | | Se-Met form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ... | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
8ETD
 
 | | Crystal Structure of Schizosaccharomyces pombe Rho1 | | Descriptor: | GTP-binding protein rho1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Huang, Q, Xie, J, Seetharaman, J. | | Deposit date: | 2022-10-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Rho1 Reveals Its Evolutionary Relationship with Other Rho GTPases.
Biology (Basel), 11, 2022
|
|
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
5N84
 
 | | TTK kinase domain in complex with Mps-BAY2b | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dual specificity protein kinase TTK, SODIUM ION, ... | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
2Y3D
 
 | | Zn-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ZINC ION | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
6WGB
 
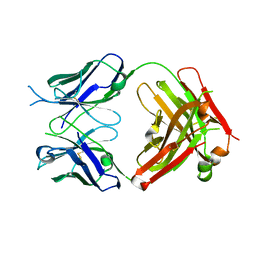 | | Crystal structure of the fab portion of dupilumab | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
