8GI8
 
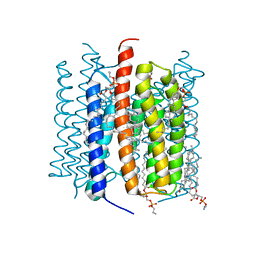 | | Kalium channelrhodopsin 1 from Hyphochytrium catenoides (HcKCR1) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Kalium Channelrhodopsin 1, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8GI9
 
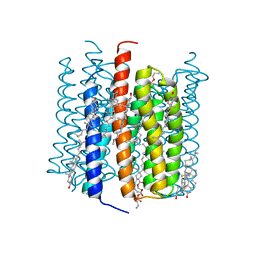 | | Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Cation Channelrhodopsin, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
4EU2
 
 | | Crystal structure of 20s proteasome with novel inhibitor K-7174 | | Descriptor: | 1,4-bis[(4E)-5-(3,4,5-trimethoxyphenyl)pent-4-en-1-yl]-1,4-diazepane, Proteasome component C1, Proteasome component C11, ... | | Authors: | Kikuchi, J, Shibayama, N, Yamada, S, Wada, T, Nobuyoshi, M, Izumi, T, Akutsu, M, Kano, Y, Ohki, M, Sugiyama, K, Park, S.-Y, Furukawa, Y. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Homopiperazine derivatives as a novel class of proteasome inhibitors with a unique mode of proteasome binding.
Plos One, 8, 2013
|
|
2IEY
 
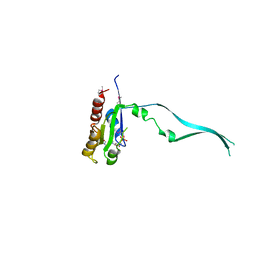 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IEZ
 
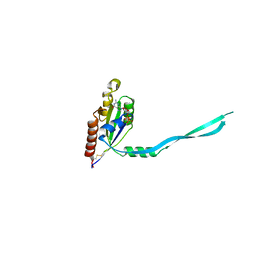 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IF0
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1DE8
 
 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1DE9
 
 | | HUMAN APE1 ENDONUCLEASE WITH BOUND ABASIC DNA AND MN2+ ION | | Descriptor: | 5'-d(*CP*TP*AP*C)-3', 5'-d(*GP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-d(P*(3DR)P*GP*AP*TP*C)-3', ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DEW
 
 | | CRYSTAL STRUCTURE OF HUMAN APE1 BOUND TO ABASIC DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', 5'-D(*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE, ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
5WKT
 
 | | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K | | Descriptor: | Rhodopsin, SULFATE ION, Transducin Galpha peptide, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
5E8D
 
 | | Crystal structure of human epiregulin in complex with the Fab fragment of murine monoclonal antibody 9E5 | | Descriptor: | CHLORIDE ION, GLYCEROL, Proepiregulin, ... | | Authors: | Kado, Y, Mizohata, E, Nagatoishi, S, Iijima, M, Shinoda, K, Miyafusa, T, Nakayama, T, Yoshizumi, T, Sugiyama, A, Kawamura, T, Lee, Y.H, Matsumura, H, Doi, H, Fujitani, H, Kodama, T, Shibasaki, Y, Tsumoto, K, Inoue, T. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-09 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Epiregulin Recognition Mechanisms by Anti-epiregulin Antibody 9E5: STRUCTURAL, FUNCTIONAL, AND MOLECULAR DYNAMICS SIMULATION ANALYSES
J.Biol.Chem., 291, 2016
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
8ELJ
 
 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
6WP8
 
 | | Proton-pumping mutant of Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | Proton-pumping rhodopsin chloride pump, RETINAL, octyl beta-D-glucopyranoside | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
8EL2
 
 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
5VRA
 
 | | 2.35-Angstrom In situ Mylar structure of human A2A adenosine receptor at 100 K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Klingel, V, Kuo, A, Kissick, D.J, Ishchenko, A, Lee, M.-Y, Xu, S, Makarov, O, Cherezov, V, Ogata, C.M, Ernst, O.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
6NWE
 
 | |
6PEL
 
 | |
6PGS
 
 | |
6PH7
 
 | | Crystal structure of bovine opsin with nerol bound | | Descriptor: | (2Z)-3,7-dimethylocta-2,6-dien-1-ol, G protein CT2 peptide, PALMITIC ACID, ... | | Authors: | Eger, B.T, Morizumi, T, Ernst, O.P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Odorant-binding site in visual opsin
To Be Published
|
|
6V51
 
 | | Spin-labeled T4 Lysozyme (9/131FnbY)-(4-Amino-TEMPO) | | Descriptor: | 4-amino-2,2,6,6-tetramethylpiperidin-1-ol, Endolysin | | Authors: | Liu, J, Morizumi, T, Ou, W.L, Wang, L, Ernst, O.P. | | Deposit date: | 2019-12-02 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Genetically Encoded Quinone Methides Enabling Rapid, Site-Specific, and Photocontrolled Protein Modification with Amine Reagents.
J.Am.Chem.Soc., 142, 2020
|
|
1OZ8
 
 | |
3PQR
 
 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
6NWD
 
 | | X-ray Crystallographic structure of Gloeobacter rhodopsin | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DECANE, DODECANE, ... | | Authors: | Ernst, O.P, Morizumi, T, Ou, W.L. | | Deposit date: | 2019-02-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Structure and Oligomerization of Gloeobacter Rhodopsin.
Sci Rep, 9, 2019
|
|
