8A9L
 
 | | Cryo-EM structure of alpha-synuclein filaments from Parkinson's disease and dementia with Lewy bodies | | Descriptor: | Alpha-synuclein, Unknown fragment | | Authors: | Yang, Y, Shi, Y, Schweighauser, M, Zhang, X.J, Kotecha, A, Murzin, A.G, Garringer, H.J, Cullinane, P, Saito, Y, Foroud, T, Warner, T.T, Hasegawa, K, Vidal, R, Murayama, S, Revesz, T, Ghetti, B, Hasegawa, M, Lashley, T, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structures of alpha-synuclein filaments from human brains with Lewy pathology.
Nature, 610, 2022
|
|
7QI4
 
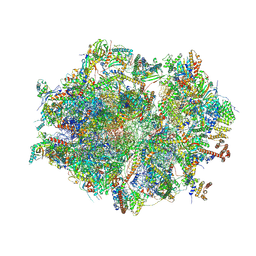 | | Human mitochondrial ribosome at 2.2 A resolution (bound to partly built tRNAs and mRNA) | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
8WUT
 
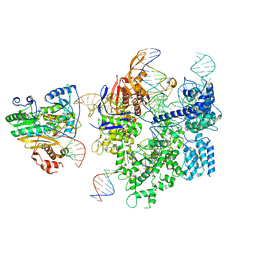 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
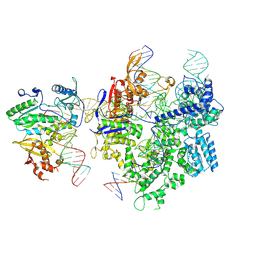 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
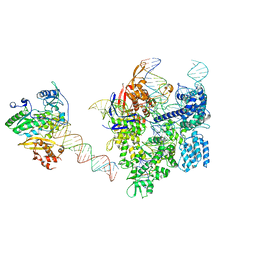 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8J12
 
 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
8J3R
 
 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
3C7X
 
 | | Hemopexin-like domain of matrix metalloproteinase 14 | | Descriptor: | CHLORIDE ION, Matrix metalloproteinase-14, SODIUM ION | | Authors: | Tochowicz, A, Itoh, Y, Maskos, K, Bode, W, Goettig, P. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions
J.Biol.Chem., 286, 2011
|
|
8QNN
 
 | | Crystal structure of a Class A beta-lactamase from Nocardia cyriacigeorgica | | Descriptor: | Beta-lactamase, CITRATE ANION | | Authors: | Feuillard, J, Couston, J, Benito, Y, Hodille, E, Dumitrescu, O, Blaise, M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical and structural characterization of a class A beta-lactamase from Nocardia cyriacigeorgica.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
6IKN
 
 | | Crystal structure of the GAS7 F-BAR domain | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
6IKO
 
 | | Crystal structure of mouse GAS7cb | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.756 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
8K1R
 
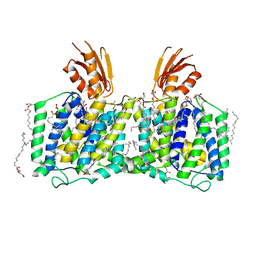 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera) | | Authors: | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | Deposit date: | 2023-07-11 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | YeeD is an essential partner for YeeE-mediated thiosulfate uptake in bacteria and regulates thiosulfate ion decomposition.
Plos Biol., 22, 2024
|
|
5H5H
 
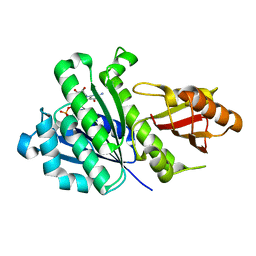 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5G
 
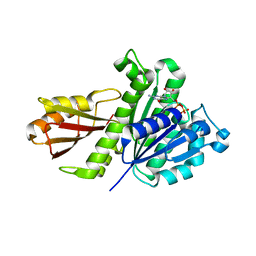 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5I
 
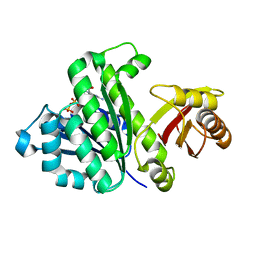 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
2YWQ
 
 | | Crystal structure of Thermus thermophilus Protein Y N-terminal domain | | Descriptor: | Ribosomal subunit interface protein | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Tatsuguchi, A, Saito, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-21 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of Thermus thermophilus Protein Y N-terminal domain
To be Published
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.384 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5Y5N
 
 | | Crystal structure of human Sirtuin 2 in complex with a selective inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]benzamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Mellini, P, Itoh, Y, Tsumoto, H, Li, Y, Suzuki, M, Tokuda, N, Kakizawa, T, Miura, Y, Takeuchi, J, Lahtela-Kakkonen, M, Suzuki, T. | | Deposit date: | 2017-08-09 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent mechanism-based sirtuin-2-selective inhibition by anin situ-generated occupant of the substrate-binding site, "selectivity pocket" and NAD+-binding site.
Chem Sci, 8, 2017
|
|
6J7C
 
 | | Crystal structure of proline racemase-like protein from Thermococcus litoralis in complex with proline | | Descriptor: | PROLINE, Proline racemase | | Authors: | Watanabe, Y, Watanabe, S, Itoh, Y, Watanabe, Y. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of substrate-bound bifunctional proline racemase/hydroxyproline epimerase from a hyperthermophilic archaeon.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
9JR2
 
 | | Cryo-EM structure of PTH-PTH1R-Gq (upright state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha-1 (miniGq), ... | | Authors: | Sano, F.K, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-09-29 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Insights into G-protein coupling preference from cryo-EM structures of Gq-bound PTH1R
To Be Published
|
|
