1UJM
 
 | |
1Y1P
 
 | | X-ray structure of aldehyde reductase with NADPH | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, Aldehyde reductase II, ... | | Authors: | Kamitori, S, Kita, K. | | Deposit date: | 2004-11-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray Structures of NADPH-dependent Carbonyl Reductase from Sporobolomyces salmonicolor Provide Insights into Stereoselective Reductions of Carbonyl Compounds
J.Mol.Biol., 352, 2005
|
|
8J82
 
 | | GaHNL-12gen (artificial S-hydroxynitrile lyase generated by GAOptimizer) | | Descriptor: | S-hydroxynitrile lyase | | Authors: | Ozawa, H, Unno, I, Sekine, R, Ito, S, Nakano, S. | | Deposit date: | 2023-04-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of evolutionary algorithm-based protein redesign method
Cell Rep Phys Sci, 5, 2024
|
|
7X7K
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Arg binding form | | Descriptor: | ARGININE, FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7J
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Lys binding form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7I
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) ligand free form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
6LU2
 
 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU3
 
 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans complexed with 4-hydroxybenzoate hydrazide | | Descriptor: | 4-oxidanylbenzohydrazide, Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU4
 
 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6M5W
 
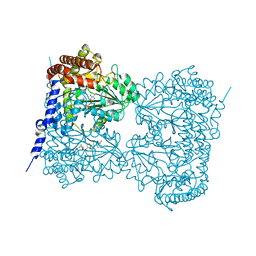 | | Co-crystal structure of human serine hydroxymethyltransferase 1 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
4KMI
 
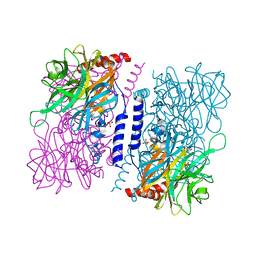 | | Crystal structure of 4-O-beta-D-mannosyl-D-glucose phosphorylase MGP complexed with PO4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-O-beta-D-mannosyl-D-glucose phosphorylase, PHOSPHATE ION | | Authors: | Nakae, S, Ito, S, Higa, M, Senoura, T, Wasaki, J, Hijikata, A, Shionyu, M, Ito, S, Shirai, T. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Novel Enzyme in Mannan Biodegradation Process 4-O-beta-d-Mannosyl-d-Glucose Phosphorylase MGP
J.Mol.Biol., 425, 2013
|
|
5XWS
 
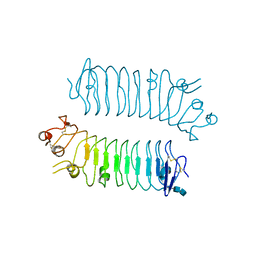 | | Crystal structure of SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat and fibronectin type-III domain-containing protein 5 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5XWU
 
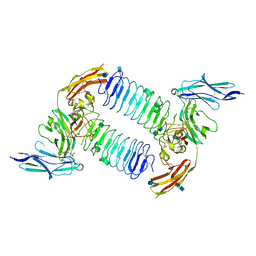 | | Crystal structure of PTPdelta Ig1-Ig3 in complex with SALM2 LRR-Ig | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5XWT
 
 | | Crystal structure of PTPdelta Ig1-Fn1 in complex with SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.178 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
2YX1
 
 | | Crystal structure of M.jannaschii tRNA m1G37 methyltransferase | | Descriptor: | Hypothetical protein MJ0883, SINEFUNGIN, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Ishii, R, Bessho, Y, Yokoyama, S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5.
Proteins, 72, 2008
|
|
2ZZN
 
 | | The complex structure of aTrm5 and tRNACys | | Descriptor: | MAGNESIUM ION, RNA (71-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Goto-Ito, S, Ito, T, Yokoyama, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2ZZM
 
 | | The complex structure of aTrm5 and tRNALeu | | Descriptor: | MAGNESIUM ION, RNA (84-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Goto-Ito, S, Ito, T, Yokoyama, S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation
Nat.Struct.Mol.Biol., 16, 2009
|
|
2YX0
 
 | | Crystal structure of P. horikoshii TYW1 | | Descriptor: | radical sam enzyme | | Authors: | Goto-Ito, S, Ishii, R, Ito, T, Shibata, R, Fusatomi, E, Sekine, S, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3AXZ
 
 | | Crystal structure of Haemophilus influenzae TrmD in complex with adenosine | | Descriptor: | ADENOSINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Yoshida, K, Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
3AY0
 
 | | Crystal structure of Methanocaldococcus jannaschii Trm5 in complex with adenosine | | Descriptor: | ADENOSINE, Uncharacterized protein MJ0883, ZINC ION | | Authors: | Goto-Ito, S, Ito, T, Hou, Y.M, Yokoyama, S. | | Deposit date: | 2011-04-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Differentiating analogous tRNA methyltransferases by fragments of the methyl donor.
Rna, 17, 2011
|
|
6M5O
 
 | | Co-crystal structure of human serine hydroxymethyltransferase 2 in complex with Pyridoxal 5'-phosphate (PLP) and glycodeoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCINE, Serine hydroxymethyltransferase, ... | | Authors: | Ota, T, Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K, Sando, S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30000663 Å) | | Cite: | Structural basis for selective inhibition of human serine hydroxymethyltransferase by secondary bile acid conjugate.
Iscience, 24, 2021
|
|
1WSD
 
 | | Alkaline M-protease form I crystal structure | | Descriptor: | CALCIUM ION, M-protease, SULFATE ION | | Authors: | Shirai, T, Suzuki, A, Yamane, T, Ashida, T, Kobayashi, T, Hitomi, J, Ito, S. | | Deposit date: | 2004-11-05 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of M-protease: phylogeny aided analysis of the high-alkaline adaptation mechanism
Protein Eng., 10, 1997
|
|
5ZQU
 
 | | Crystal structure of tetrameric RXRalpha-LBD complexed with partial agonist CBt-PMN | | Descriptor: | 1-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)benzotriazole-5-carboxylic acid, BROMIDE ION, Retinoic acid receptor RXR-alpha | | Authors: | Miyashita, Y, Numoto, N, Arulmozhiraja, S, Nakano, S, Matsuo, N, Shimizu, K, Kakuta, H, Ito, S, Ikura, T, Ito, N, Tokiwa, H. | | Deposit date: | 2018-04-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.60038781 Å) | | Cite: | Dual conformation of the ligand induces the partial agonistic activity of retinoid X receptor alpha (RXR alpha ).
FEBS Lett., 593, 2019
|
|
3VRH
 
 | | Crystal structure of ph0300 | | Descriptor: | BICINE, Putative uncharacterized protein PH0300, ZINC ION | | Authors: | Nakagawa, H, Kuratani, M, Goto-Ito, S, Ito, T, Sekine, S.I, Yokoyama, S. | | Deposit date: | 2012-04-10 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA.
Proteins, 2013
|
|
1WKY
 
 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
