6QPZ
 
 | | Crystal structure of as isolated Y323E mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
5DS1
 
 | | Core domain of the class II small heat-shock protein HSP 17.7 from Pisum sativum | | Descriptor: | 17.1 kDa class II heat shock protein | | Authors: | Hochberg, G.K.A, Laganoswky, A, Allison, T.A, Shepherd, D.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
4NH2
 
 | | Crystal structure of AmtB from E. coli bound to phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Ammonia channel | | Authors: | Laganowsky, A, Reading, E, Allison, T.M, Robinson, C.V. | | Deposit date: | 2013-11-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membrane proteins bind lipids selectively to modulate their structure and function.
Nature, 510, 2014
|
|
5G2N
 
 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
5DS2
 
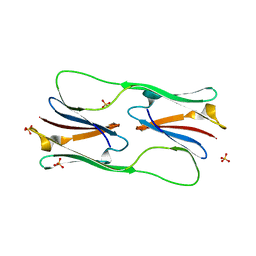 | | Core domain of the class I small heat-shock protein HSP 18.1 from Pisum sativum | | Descriptor: | 18.1 kDa class I heat shock protein, SULFATE ION | | Authors: | Shepherd, D.A, Laganowsky, A, Allison, T.M, Hochberg, G.K.A, Benesch, J.L.P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions.
Science, 359, 2018
|
|
6AZ3
 
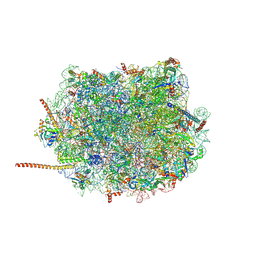 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
4R07
 
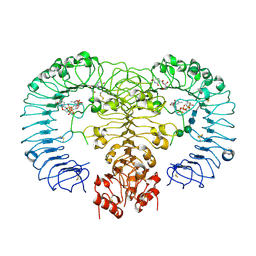 | | Crystal structure of human TLR8 in complex with ORN06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3'-O-[(R)-{[(2R,3aR,4R,6R,6aR)-6-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-hydroxy-2-oxidotetrahydrofuro[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]uridine 5'-(dihydrogen phosphate), ... | | Authors: | Tanji, H, Ohto, U, Shibata, T, Taoka, M, Yamauchi, Y, Isobe, T, Miyake, K, Shimizu, T. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toll-like receptor 8 senses degradation products of single-stranded RNA
Nat.Struct.Mol.Biol., 22, 2015
|
|
5EXG
 
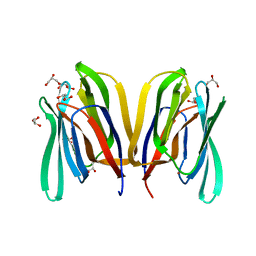 | |
4IXX
 
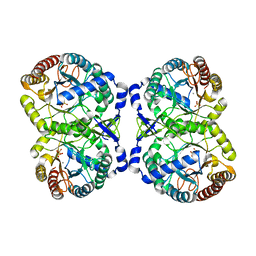 | | Crystal structure of S213G variant DAH7PS without Tyr bound from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, SULFATE ION | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
3NAX
 
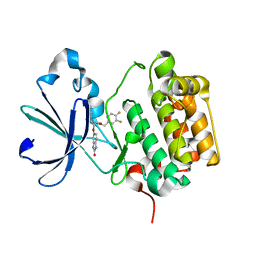 | | PDK1 in complex with inhibitor MP7 | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor.
J.Biol.Chem., 286, 2011
|
|
3NAY
 
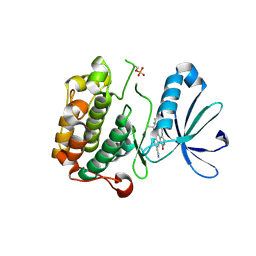 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
3NUT
 
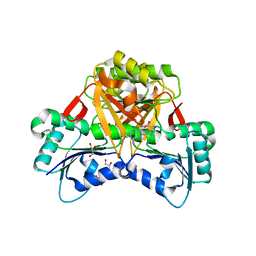 | |
3QKL
 
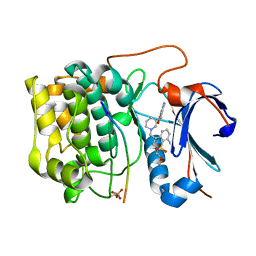 | | Spirochromane Akt Inhibitors | | Descriptor: | GSK-3 beta peptide, N-{(2S)-3-[(3S)-8',9'-dihydro-1H,3'H-spiro[piperidine-3,7'-pyrano[3,2-e]indazol]-1-yl]-2-hydroxypropyl}-N-(2-ethoxyethyl)-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QKK
 
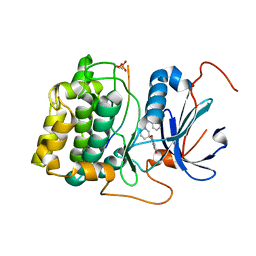 | | Spirochromane Akt Inhibitors | | Descriptor: | Glycogen synthase kinase-3 beta, N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(2R)-6-hydroxy-4-oxo-3,4-dihydro-1'H-spiro[chromene-2,3'-piperidin]-1'-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Kallan, N.C, Spencer, K.L, Blake, J.F, Xu, R, Heizer, J, Bencsik, J.R, Mitchell, I.S, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and SAR of spirochromane Akt inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q6U
 
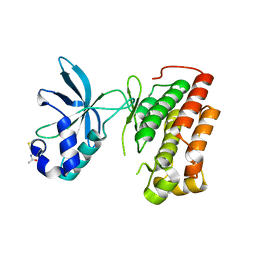 | | Structure of the apo MET receptor kinase in the dually-phosphorylated, activated state | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Allison, T, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
3QKM
 
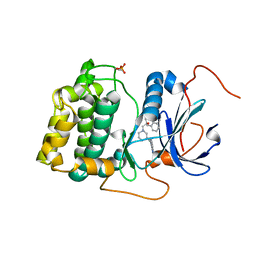 | | Spirocyclic sulfonamides as AKT inhibitors | | Descriptor: | N-(2-ethoxyethyl)-N-{(2S)-2-hydroxy-3-[(5R)-2-(quinazolin-4-yl)-2,7-diazaspiro[4.5]dec-7-yl]propyl}-2,6-dimethylbenzenesulfonamide, RAC-alpha serine/threonine-protein kinase | | Authors: | Xu, R, Banka, A, Blake, J.F, Mitchell, I.S, Wallace, E.M, Gloor, S.L, Martinson, M, Risom, T, Gross, S.D, Morales, T, Vigers, G.P.A, Brandhuber, B.J, Skelton, N.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of spirocyclic sulfonamides as potent Akt inhibitors with exquisite selectivity against PKA.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3B0W
 
 | | Crystal structure of the orphan nuclear receptor ROR(gamma)t ligand-binding domain in complex with digoxin | | Descriptor: | DIGOXIN, Nuclear receptor ROR-gamma | | Authors: | Fujita-Sato, S, Ito, S, Isobe, T, Ohyama, T, Wakabayashi, K, Morishita, K, Ando, O, Isono, F. | | Deposit date: | 2011-06-17 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Digoxin That Antagonizes ROR{gamma}t Receptor Activity and Suppresses Th17 Cell Differentiation and Interleukin (IL)-17 Production
J.Biol.Chem., 286, 2011
|
|
2BM2
 
 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
1A63
 
 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
1HNB
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1HNC
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS MU GLUTATHIONE TRANSFERASE GSTM2-2: EFFECTS OF LATTICE PACKING ON CONFORMATIONAL HETEROGENEITY | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Raghunathan, S, Chandross, R.J, Kretsinger, R.H, Allison, T.J, Penington, C.J, Rule, G.S. | | Deposit date: | 1993-10-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human class mu glutathione transferase GSTM2-2. Effects of lattice packing on conformational heterogeneity.
J.Mol.Biol., 238, 1994
|
|
1YPY
 
 | | Crystal Structure of Vaccinia Virus L1 protein | | Descriptor: | Virion membrane protein | | Authors: | Su, H.P, Garman, S.C, Allison, T.J, Fogg, C, Moss, B, Garboczi, D.N. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The 1.51-Angstrom structure of the poxvirus L1 protein, a target of potent neutralizing antibodies.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4HSO
 
 | | Crystal structure of S213G variant DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
4HSN
 
 | | Crystal structure of DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
