3VH4
 
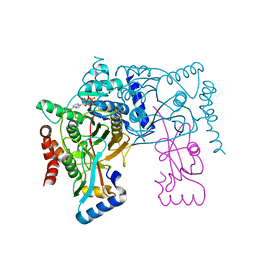 | | Crystal structure of Atg7CTD-Atg8-MgATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Autophagy-related protein 8, MAGNESIUM ION, ... | | Authors: | Noda, N.N, Satoo, K, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VU0
 
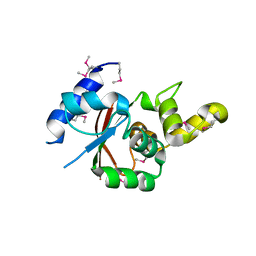 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VP7
 
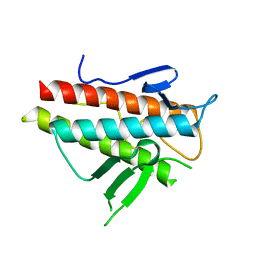 | | Crystal structure of the beta-alpha repeated, autophagy-specific (BARA) domain of Vps30/Atg6 | | Descriptor: | Vacuolar protein sorting-associated protein 30 | | Authors: | Noda, N.N, Kobayashi, T, Adachi, W, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy.
J.Biol.Chem., 287, 2012
|
|
3VH2
 
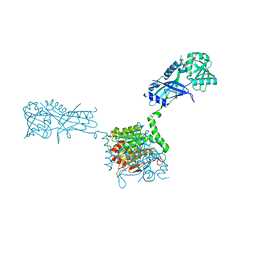 | |
3VGO
 
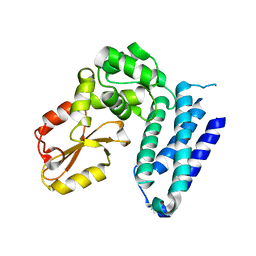 | |
3VU1
 
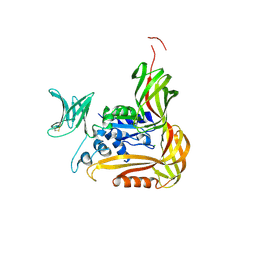 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VX6
 
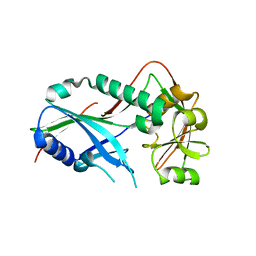 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VXW
 
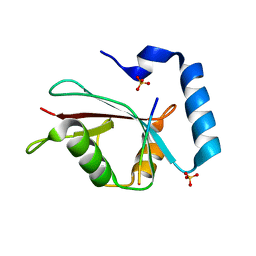 | |
3VX7
 
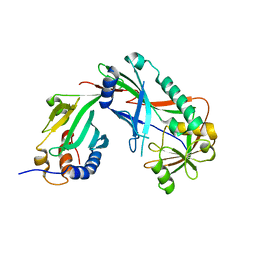 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3W1S
 
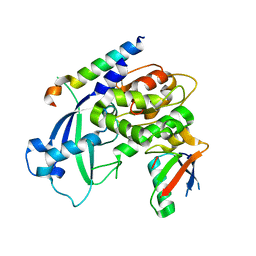 | | Crystal structure of Saccharomyces cerevisiae Atg12-Atg5 conjugate bound to the N-terminal domain of Atg16 | | Descriptor: | Autophagy protein 16, Autophagy protein 5, Ubiquitin-like protein ATG12 | | Authors: | Noda, N.N, Fujioka, Y, Hanada, T, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Embo Rep., 14, 2013
|
|
3VH1
 
 | |
3VQI
 
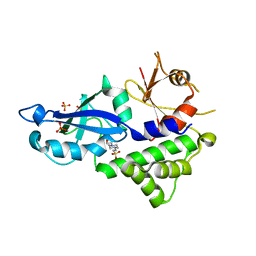 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
1UCF
 
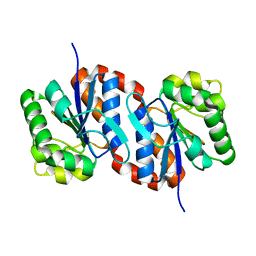 | | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Honbou, K, Suzuki, N.N, Horiuchi, M, Niki, T, Taira, T, Ariga, H, Inagaki, F. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
J.BIOL.CHEM., 278, 2003
|
|
1UEC
 
 | | Crystal structure of autoinhibited form of tandem SH3 domain of p47phox | | Descriptor: | Neutrophil cytosol factor 1 | | Authors: | Yuzawa, S, Suzuki, N.N, Fujioka, Y, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-05-11 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A molecular mechanism for autoinhibition of the tandem SH3 domains of p47phox, the regulatory subunit of the phagocyte NADPH oxidase
Genes Cells, 9, 2004
|
|
1TZ1
 
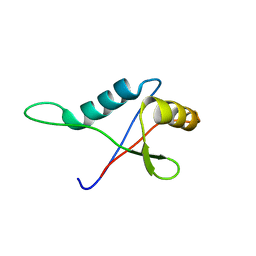 | | Solution structure of the PB1 domain of CDC24P (short form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Terasawa, H, Ogura, K, Noda, Y, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PB1 domain of CDC24P (short form)
To be Published
|
|
1TH5
 
 | | Solution structure of C-terminal domain of NifU-like protein from Oryza sativa | | Descriptor: | NifU1 | | Authors: | Kumeta, H, Ogura, K, Asayama, M, Katoh, S, Katoh, E, Inagaki, F. | | Deposit date: | 2004-06-01 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the domain II of a chloroplastic NifU-like protein OsNifU1A.
J.Biomol.Nmr, 38, 2007
|
|
2DYM
 
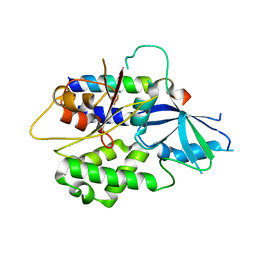 | |
2DYT
 
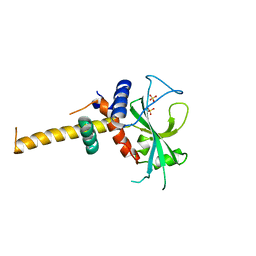 | |
2DYO
 
 | |
2CY7
 
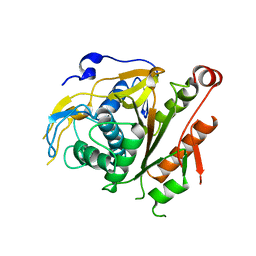 | | The crystal structure of human Atg4B | | Descriptor: | Cysteine protease APG4B | | Authors: | Sugawara, K, Suzuki, N.N, Fujioka, Y, Mizushima, N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2005-07-05 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Specificity and Catalysis of Human Atg4B Responsible for Mammalian Autophagy
J.Biol.Chem., 280, 2005
|
|
2CJN
 
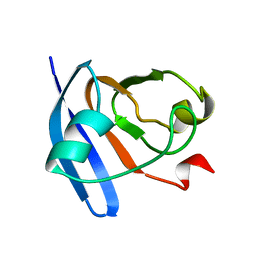 | | STRUCTURE OF FERREDOXIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
2CJO
 
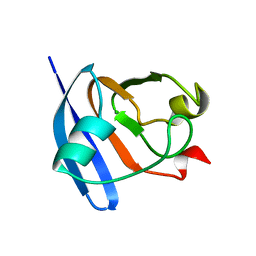 | | STRUCTURE OF FERREDOXIN, NMR, 10 STRUCTURES | | Descriptor: | FERREDOXIN | | Authors: | Hatanaka, H, Tanimura, R, Katoh, S, Inagaki, F. | | Deposit date: | 1997-02-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ferredoxin from the thermophilic cyanobacterium Synechococcus elongatus and its thermostability.
J.Mol.Biol., 268, 1997
|
|
2CZO
 
 | | Solution Structure of the PX Domain of Bem1p | | Descriptor: | Bud emergence protein 1 | | Authors: | Maeda, A, Ogura, K, Horiuchi, M, Kumeta, H, Fujioka, Y, Inagaki, F. | | Deposit date: | 2005-07-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PX domain of Bem1p
To be Published
|
|
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
2EYZ
 
 | | CT10-Regulated Kinase isoform II | | Descriptor: | v-crk sarcoma virus CT10 oncogene homolog isoform a | | Authors: | Kobashigawa, Y, Tanaka, S, Inagaki, F. | | Deposit date: | 2005-11-10 | | Release date: | 2006-11-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
