7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6GB5
 
 | | Structure of H-2Db with truncated SEV peptide and GL | | Descriptor: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3BVB
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
3BVF
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BPM
 
 | | Crystal Structure of Falcipain-3 with Its inhibitor, Leupeptin | | Descriptor: | Cysteine protease falcipain-3, Leupeptin, SULFATE ION | | Authors: | kerr, I.D, Lee, J.H, Brinen, L.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity.
J.Med.Chem., 52, 2009
|
|
3BVI
 
 | | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin | | Descriptor: | FE (III) ION, Ferritin, GLYCEROL | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Shin, H.J, Yang, I.S. | | Deposit date: | 2008-01-07 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the iron uptake mechanism of Helicobacter pylori ferritin
To be Published
|
|
3BVA
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | Descriptor: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
4DP6
 
 | | The 1.67 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DPC
 
 | | The 1.06 Angstrom crystal structure of reduced (CuI) poplar plastocyanin A at pH 8.0 | | Descriptor: | COPPER (I) ION, Plastocyanin A, chloroplastic | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
3BPF
 
 | | Crystal Structure of Falcipain-2 with Its inhibitor, E64 | | Descriptor: | Cysteine protease falcipain-2, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | kerr, I.D, Lee, J.H, Brinen, L.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of falcipain-2 and falcipain-3 bound to small molecule inhibitors: implications for substrate specificity.
J.Med.Chem., 52, 2009
|
|
1GPI
 
 | |
4UXS
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
3C2A
 
 | | Antibody Fab fragment 447-52D in complex with UG1033 peptide | | Descriptor: | Envelope glycoprotein, Fab 447-52D heavy chain, Fab 447-52D light chain | | Authors: | Dhillon, A.K, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2008-01-24 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3BP8
 
 | | Crystal structure of Mlc/EIIB complex | | Descriptor: | ACETATE ION, PTS system glucose-specific EIICB component, Putative NAGC-like transcriptional regulator, ... | | Authors: | An, Y.J, Jung, H.I, Cha, S.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
1HB1
 
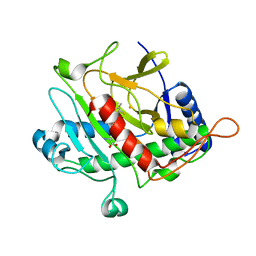 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC ACOV FE COMPLEX) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHASE, N6-[(1R)-2-{[(1R)-1-CARBOXY-2-METHYLPROPYL]OXY}-1-(MERCAPTOMETHYL)-2-OXOETHYL]-6-OXO-D-LYSINE, ... | | Authors: | Ogle, J.M, Clifton, I.J, Rutledge, P.J, Elkins, J.M, Burzlaff, N.I, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Alternative Oxidation by Isopenicillin N Synthase Observed by X-Ray Diffraction.
Chem.Biol., 8, 2001
|
|
5C4X
 
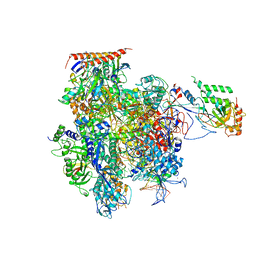 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
8C5P
 
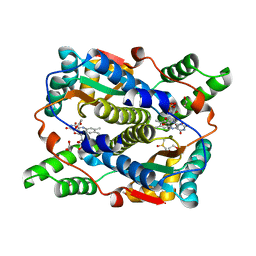 | | E. coli NfsB mutant N71S T41L with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Day, M.A, White, S.A, Hyde, E.I. | | Deposit date: | 2023-01-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8CJ0
 
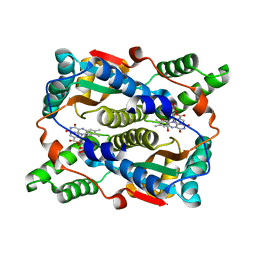 | | E. coli NfsB-T41Q/N71S/F124T/M127V mutant bound to nicotinate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | Authors: | White, S.A, Hyde, E.I, Day, M.A. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8C5E
 
 | | E. coli NfsB-T41Q/N71S/F124T mutant bound to nicotinic acid | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | Authors: | White, S.A, Hyde, E.I, Day, M.A. | | Deposit date: | 2023-01-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8C5F
 
 | | E. coli NfsB-T41Q/N71S/F124T mutant bound to acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | White, S.A, Hyde, E.I, Day, M.A. | | Deposit date: | 2023-01-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
1GND
 
 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Authors: | Schalk, I, Zeng, K, Wu, S.-K, Stura, E.A, Metteson, J, Huang, M, Tandon, A, Wilson, I.A, Balch, W.E. | | Deposit date: | 1996-07-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and mutational analysis of Rab GDP-dissociation inhibitor.
Nature, 381, 1996
|
|
7OCC
 
 | | NTD of resting state GluA1/A2 heterotertramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 1, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
8CO6
 
 | | Subtomogram average of Immature Rotavirus TLP penton | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8CIM
 
 | | BA.2-07 FAB IN COMPLEX WITH SARS-COV-2 BA.2.12.1 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.2-07 FAB HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Isolation of a pair of potent broadly neutralizing mAb binding to RBD and SD1 domains of SARS-CoV-2
Res Sq, 2023
|
|
