6CI4
 
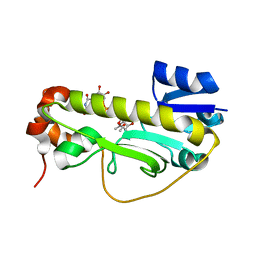 | | Crystal structure of the formyltransferase PseJ from Anoxybacillus kamchatkensis soaked with UDP-4-amino-4,6-dideoxy-L-AltNAc | | Descriptor: | (2R,3R,4S,5R,6S)-3-(acetylamino)-5-amino-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, formyltransferase PseJ | | Authors: | Harb, I, Reimer, J.M, Schmeing, T.M. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.824068 Å) | | Cite: | Structural Insight into a Novel Formyltransferase and Evolution to a Nonribosomal Peptide Synthetase Tailoring Domain.
ACS Chem. Biol., 13, 2018
|
|
6K8M
 
 | |
1OB1
 
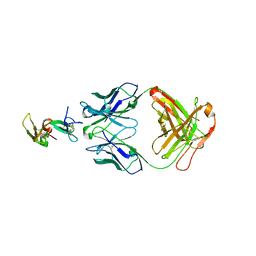 | | Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 | | Descriptor: | ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pizarro, J.C, Chitarra, V, Verger, D, Holm, I, Petres, S, Dartville, S, Nato, F, Longacre, S, Bentley, G.A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
J.Mol.Biol., 328, 2003
|
|
6KEC
 
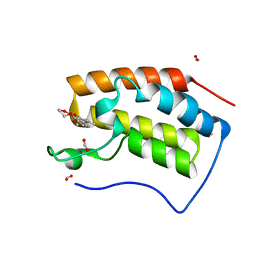 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
1KMD
 
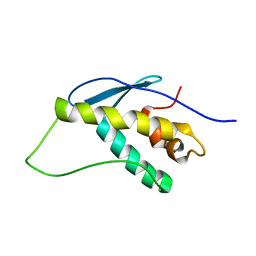 | | SOLUTION STRUCTURE OF THE VAM7P PX DOMAIN | | Descriptor: | Vacuolar morphogenesis protein VAM7 | | Authors: | Lu, J, Garcia, J, Dulubova, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Vam7p PX domain.
Biochemistry, 41, 2002
|
|
6MK5
 
 | |
1KPQ
 
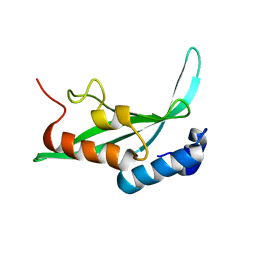 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
2CH0
 
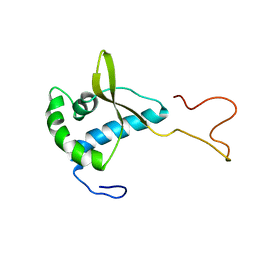 | | Solution structure of the human MAN1 C-terminal domain (residues 655- 775) | | Descriptor: | INNER NUCLEAR MEMBRANE PROTEIN MAN1 | | Authors: | Caputo, S, Couprie, J, Duband-Goulet, I, Lin, F, Braud, S, Gondry, M, Worman, H.J, Gilquin, B, Zinn-Justin, S. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain.
J. Biol. Chem., 281, 2006
|
|
1KVM
 
 | | X-ray Crystal Structure of AmpC WT beta-Lactamase in Complex with Covalently Bound Cephalothin | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
1KWJ
 
 | | solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans, minimized average structure | | Descriptor: | HEME C, cytochrome c7 | | Authors: | Assfalg, M, Bertini, I, Turano, P, Bruschi, M, Durand, M.C, Giudici-Orticoni, M.T, Dolla, A. | | Deposit date: | 2002-01-29 | | Release date: | 2002-02-06 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A quick solution structure determination of the fully oxidized double mutant K9-10A cytochrome c7 from Desulfuromonas acetoxidans and mechanistic implications.
J.Biomol.NMR, 22, 2002
|
|
2CLF
 
 | | Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2- amino-1-ethylphosphate (F6) - highF6 complex | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
1YXJ
 
 | | Crystal structure of human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) at low pH | | Descriptor: | 1,2-ETHANEDIOL, oxidised low density lipoprotein (lectin-like) receptor 1 | | Authors: | Ohki, I, Ishigaki, T, Oyama, T, Matsunaga, S, Xie, Q, Ohnishi-Kameyama, M, Murata, T, Tsuchiya, D, Machida, S, Morikawa, K, Tate, S. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human lectin-like, oxidized low-density lipoprotein receptor 1 ligand binding domain and its ligand recognition mode to OxLDL.
Structure, 13, 2005
|
|
1Z64
 
 | | NMR Solution Structure of Pleurocidin in DPC Micelles | | Descriptor: | Pleruocidin | | Authors: | Syvitski, R.T, Burton, I, Mattatall, N.R, Douglas, S.E, Jakeman, D.L. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the antimicrobial peptide pleurocidin from winter flounder.
Biochemistry, 44, 2005
|
|
1ZA0
 
 | | X-ray structure of putative acyl-ACP desaturase DesA2 from Mycobacterium tuberculosis H37Rv | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, POSSIBLE ACYL-[ACYL-CARRIER PROTEIN] DESATURASE DESA2 (ACYL-[ACP] DESATURASE) (STEAROYL-ACP DESATURASE) | | Authors: | Dyer, H.D, Lyle, K.S, Rayment, I, Fox, B.G. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of putative acyl-ACP desaturase DesA2 from Mycobacterium tuberculosis H37Rv.
Protein Sci., 14, 2005
|
|
1NPK
 
 | | REFINED X-RAY STRUCTURE OF DICTYOSTELIUM NUCLEOSIDE DIPHOSPHATE KINASE AT 1,8 ANGSTROMS RESOLUTION | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Lebras, G, Lascu, I, Veron, M. | | Deposit date: | 1994-07-27 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined X-ray structure of Dictyostelium discoideum nucleoside diphosphate kinase at 1.8 A resolution.
J.Mol.Biol., 243, 1994
|
|
1ZJC
 
 | | Aminopeptidase S from S. aureus | | Descriptor: | COBALT (II) ION, aminopeptidase ampS | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Staphylococcus aureus Aminopeptidase S Is a Founding Member of a New Peptidase Clan.
J.Biol.Chem., 280, 2005
|
|
1ZK6
 
 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
3RFM
 
 | | Thermostabilised adenosine A2A receptor in complex with caffeine | | Descriptor: | Adenosine receptor A2a, CAFFEINE | | Authors: | Dore, A.S, Robertson, N, Errey, J.C, Ng, I, Hollenstein, K, Tehan, B, Hurrell, E, Bennett, K, Congreve, M, Magnani, F, Tate, C.G, Weir, M, Marshall, F.H. | | Deposit date: | 2011-04-06 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine
Structure, 19, 2011
|
|
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
6NIT
 
 | |
1ZPL
 
 | | E. coli F17a-G lectin domain complex with GlcNAc(beta1-O)Me | | Descriptor: | F17a-G, methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Buts, L, Wellens, A, Van Molle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2005-05-17 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of natural variation in bacterial F17G adhesins on crystallization behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NOR
 
 | | TWO-DIMENSIONAL 1H-NMR STUDY OF THE SPATIAL STRUCTURE OF NEUROTOXIN II FROM NAJA OXIANA | | Descriptor: | NEUROTOXIN II | | Authors: | Golovanov, A.P, Utkin, Y.N, Lomize, A.L, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 1993-04-05 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Eur.J.Biochem., 213, 1993
|
|
1NPD
 
 | | X-RAY STRUCTURE OF SHIKIMATE DEHYDROGENASE COMPLEXED WITH NAD+ FROM E.COLI (YDIB) NORTHEAST STRUCTURAL GENOMICS RESEARCH CONSORTIUM (NESG) TARGET ER24 | | Descriptor: | HYPOTHETICAL SHIKIMATE 5-DEHYDROGENASE-LIKE PROTEIN YDIB, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Kuzin, A.P, Lee, I, Rost, B, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 2.3-A crystal structure of the shikimate 5-dehydrogenase orthologue YdiB from Escherichia coli suggests a novel catalytic environment for an NAD-dependent dehydrogenase
J.Biol.Chem., 278, 2003
|
|
1NPZ
 
 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Seddon, A.P, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
