6X1S
 
 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
6O3G
 
 | |
6NQP
 
 | | Crystal structure of fast switching M159T mutant of fluorescent protein Dronpa (Dronpa2), Y63(2,3-F2Y) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Electrostatic control of photoisomerization pathways in proteins.
Science, 367, 2020
|
|
6Z3K
 
 | | Structure of protective antibody 38-1-10A Fab | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain, Light Chain | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6ZG3
 
 | | the structure of ECF PanT transporter in a complex with a nanobody | | Descriptor: | CA14381 nanobody, CITRIC ACID, Conserved hypothetical membrane protein, ... | | Authors: | Setyawati, I, Guskov, A, Slotboom, D.J. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In vitro reconstitution of dynamically interacting integral membrane subunits of energy-coupling factor transporters.
Elife, 9, 2020
|
|
6NQQ
 
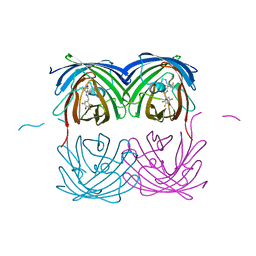 | | Crystal structure of fast switching M159T mutant of fluorescent protein Dronpa (Dronpa2), Y63(2,3,5-F3Y) | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrostatic control of photoisomerization pathways in proteins.
Science, 367, 2020
|
|
6NMP
 
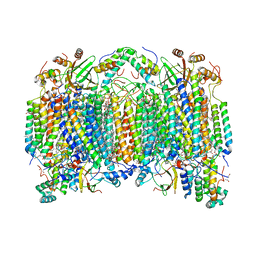 | | SFX structure of oxidized cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8ANQ
 
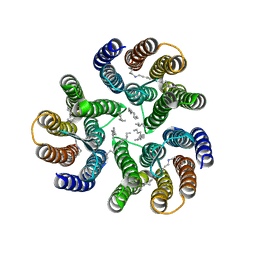 | | Crystal structure of the microbial rhodopsin from Sphingomonas paucimobilis (SpaR) | | Descriptor: | Bacteriorhodopsin, EICOSANE | | Authors: | Kovalev, K, Okhrimenko, I, Marin, E, Gordeliy, V. | | Deposit date: | 2022-08-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mirror proteorhodopsins.
Commun Chem, 6, 2023
|
|
6O3U
 
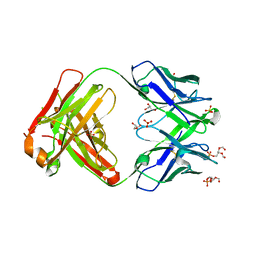 | |
6O3J
 
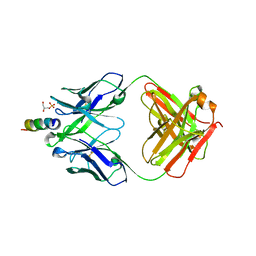 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6ZAK
 
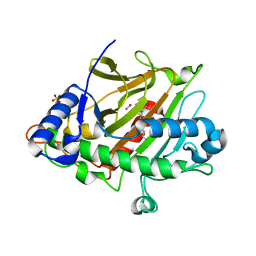 | | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV. | | Descriptor: | Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, SULFATE ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
To Be Published
|
|
7OYA
 
 | | Cryo-EM structure of the 1 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYB
 
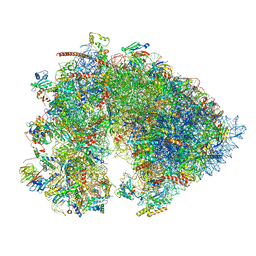 | | Cryo-EM structure of the 6 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
2C7U
 
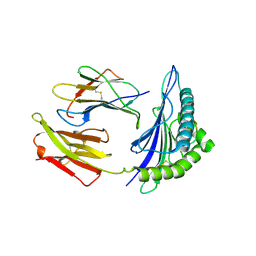 | | Conflicting selective forces affect CD8 T-cell receptor contact sites in an HLA-A2 immunodominant HIV epitope. | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Iversen, A.K, Stewart-Jones, G, Learn, G.H, Christie, N, Sylvester-Hviid, C, Armitage, A.E, Kaul, R, Beattie, T, Lee, J.K, Li, Y, Chotiyarnwong, P, Dong, T, Xu, X, Luscher, M.A, MacDonald, K, Ullum, H, Klarlund-Pedersen, B, Skinhoj, P, Fugger, J.L, Buus, S, Mullins, J.I, Jones, E.Y, van der Merwe, P.A, McMichael, A.J. | | Deposit date: | 2005-11-29 | | Release date: | 2006-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conflicting Selective Forces Affect T Cell Receptor Contacts in an Immunodominant Human Immunodeficiency Virus Epitope.
Nat.Immunol., 7, 2006
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7Q4M
 
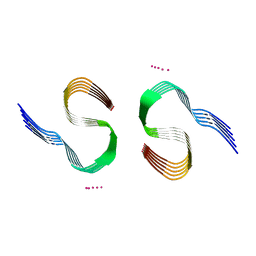 | | Type II beta-amyloid 42 Filaments from Human Brain | | Descriptor: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | Authors: | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2021-11-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
7Q5R
 
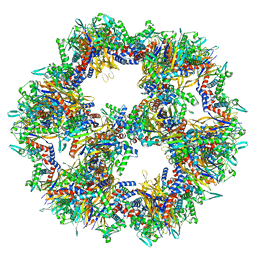 | | Protein community member pyruvate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Acetyltransferase component of pyruvate dehydrogenase complex | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5Q
 
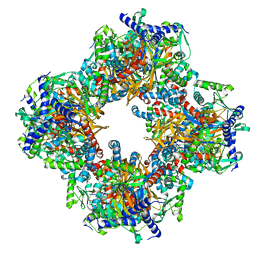 | | Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
7Q5S
 
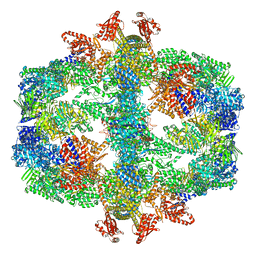 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
6WH9
 
 | | Ketoreductase from module 1 of the 6-deoxyerythronolide B synthase (KR1) in complex with antibody fragment (Fab) 1D10 | | Descriptor: | 1D10 (Fab heavy chain), 1D10 (Fab light chain), KR1, ... | | Authors: | Cogan, D.P, Mathews, I.I, Khosla, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Antibody Probes of Module 1 of the 6-Deoxyerythronolide B Synthase Reveal an Extended Conformation During Ketoreduction.
J.Am.Chem.Soc., 142, 2020
|
|
6NQL
 
 | | Crystal structure of fast switching M159T mutant of fluorescent protein Dronpa (Dronpa2), Y63(3-ClY) | | Descriptor: | Fluorescent protein Dronpa, TRIETHYLENE GLYCOL | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Electrostatic control of photoisomerization pathways in proteins.
Science, 367, 2020
|
|
6GB6
 
 | | Structure of H-2Kb with dipeptide GL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ZZ8
 
 | | MB_CRS6-15 bound to CrSAS-6_6HR | | Descriptor: | Centriole protein, Protein B | | Authors: | Hatzopoulos, G.N, Kukenshoner, T, Banterle, N, Favez, T, Fluckiger, I, Hantschel, O, Gonczy, P. | | Deposit date: | 2020-08-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly.
Nat Commun, 12, 2021
|
|
