4WE3
 
 | | STRUCTURE OF THE BINARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP MONOCLINIC CRYSTAL FORM | | Descriptor: | Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-09 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
6PUY
 
 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
4U8K
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Q107A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
2C61
 
 | | Crystal structure of the non-catalytic B subunit of A-type ATPase from M. mazei Go1 | | Descriptor: | A-TYPE ATP SYNTHASE NON-CATALYTIC SUBUNIT B | | Authors: | Schaefer, I, Bailer, S.M, Dueser, M, Boersch, M, Bernal, R.A, Grueber, G, Stock, D. | | Deposit date: | 2005-11-06 | | Release date: | 2006-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Archaeal A1Ao ATP Synthase Subunit B from Methanosarcina Mazei Go1: Implications of Nucleotide-Binding Differences in the Major A1Ao Subunits a and B. Subunits a and B
J.Mol.Biol., 358, 2006
|
|
1INJ
 
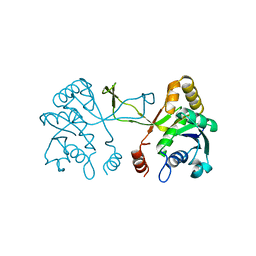 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
3LN8
 
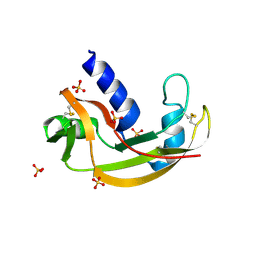 | | The X-ray structure of Zf-RNase-1 from a new crystal form at pH 7.3 | | Descriptor: | HYDROLASE, SULFATE ION | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-02-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
1IF9
 
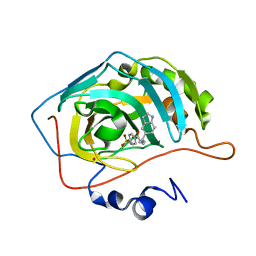 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IQI
 
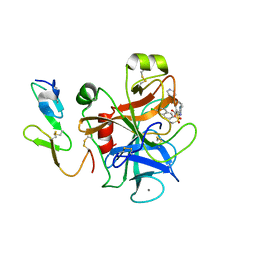 | | Human coagulation factor Xa in complex with M55125 | | Descriptor: | 4-[(6-CHLORO-2-NAPHTHALENYL)SULFONYL]-1-[[1-(4-PYRIDINYL)-4-PIPERIDINYL] METHYL]-2-PIPERAZINECARBOXYLIC ACID, CALCIUM ION, coagulation Factor Xa | | Authors: | Shiromizu, I, Matsusue, T. | | Deposit date: | 2001-07-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Factor Xa Specific Inhibitor that Induces the Novel Binding Model in Complex with Human Fxa
To be Published
|
|
3UZA
 
 | | Thermostabilised Adenosine A2A receptor in complex with 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine | | Descriptor: | 6-(2,6-dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine, Adenosine receptor A2a | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Tehan, B, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
6PW4
 
 | | Cryo-EM Structure of Thermo-Sensitive TRP Channel TRP1 from the Alga Chlamydomonas reinhardtii in Detergent | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | McGoldrick, L.L, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the thermo-sensitive TRP channel TRP1 from the alga Chlamydomonas reinhardtii.
Nat Commun, 10, 2019
|
|
6PXE
 
 | |
4WDR
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
5VYK
 
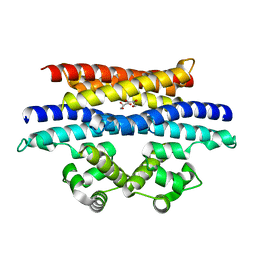 | | Crystal structure of the BRS domain of BRAF in complex with the CC-SAM domain of KSR1 | | Descriptor: | Chimera protein of BRS domain of BRAF and CC-SAM domain of KSR1,Serine/threonine-protein kinase B-raf, GLYCEROL | | Authors: | Maisonneuve, P, Kurinov, I, Marullo, S.A, Lavoie, H, Thevakumaran, N, Sahmi, M, Jin, T, Therrien, M, SIcheri, F. | | Deposit date: | 2017-05-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | MEK drives BRAF activation through allosteric control of KSR proteins.
Nature, 554, 2018
|
|
6Q20
 
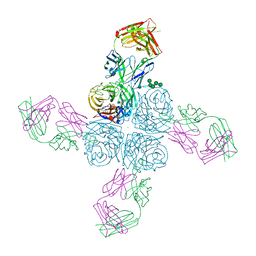 | |
4U8J
 
 | | Structure of Aspergillus fumigatus UDP-Galactopyranose mutase mutant Y104A | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Qureshi, I.A, Chaudhary, R, Tanner, J.J. | | Deposit date: | 2014-08-03 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Contributions of Unique Active Site Residues of Eukaryotic UDP-Galactopyranose Mutases to Substrate Recognition and Active Site Dynamics.
Biochemistry, 53, 2014
|
|
4UUI
 
 | |
5VYB
 
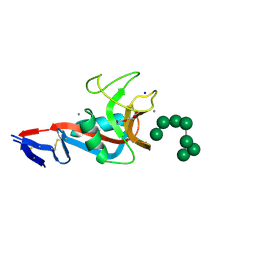 | | Structure of the carbohydrate recognition domain of Dectin-2 complexed with a mammalian-type high mannose Man9GlcNAc2 oligosaccharide | | Descriptor: | C-type lectin domain family 6 member A, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rex, M.J, Drickamer, K, Taylor, M.E, Weis, W.I. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of pathogen recognition by human dectin-2.
J. Biol. Chem., 292, 2017
|
|
8QWD
 
 | | Apo ReChb | | Descriptor: | MAGNESIUM ION, ReChb | | Authors: | Lopez-Alonso, J.P, Ubarretxena-Belandia, I, Tascon, I. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection.
Nat.Biotechnol., 2024
|
|
2BXF
 
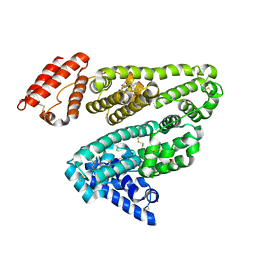 | | Human serum albumin complexed with diazepam | | Descriptor: | 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BFR
 
 | | The Macro domain is an ADP-ribose binding module | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL PROTEIN AF1521, MAGNESIUM ION | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
1IU8
 
 | | The X-ray Crystal Structure of Pyrrolidone-Carboxylate Peptidase from Hyperthermophilic Archaeon Pyrococcus horikoshii | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Sokabe, M, Kawamura, T, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The X-ray crystal structure of pyrrolidone-carboxylate peptidase from hyperthermophilic archaea Pyrococcus horikoshii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1IVT
 
 | | NMR structures of the C-terminal globular domain of human lamin A/C | | Descriptor: | Lamin A/C | | Authors: | Krimm, I, Ostlund, C, Gilquin, B, Couprie, J, Hossenlopp, P, Mornon, J.P, Bonn, G, Courvalin, J.C, Worman, H.J, Zinn-Justin, S. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy.
Structure, 10, 2002
|
|
2BFQ
 
 | | MACRO DOMAINS ARE ADP-RIBOSE BINDING MOLECULES | | Descriptor: | HYPOTHETICAL PROTEIN AF1521, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
1SQ7
 
 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-18 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
5W3Q
 
 | | L28F E.coli DHFR in complex with NADPH | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Oyen, D, Wright, P.E, Wilson, I.A. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
