8AIH
 
 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | Descriptor: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8Q6Q
 
 | |
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
8Q65
 
 | |
6KYP
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-clofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-09-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8AII
 
 | | High Resolution Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase Complexed with Adenine | | Descriptor: | ADENINE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
5G4X
 
 | | The crystal structure of the SHANK3 N-terminus | | Descriptor: | 1,2-ETHANEDIOL, SH3 AND MULTIPLE ANKYRIN REPEAT DOMAINS PROTEIN 3 | | Authors: | Zacharchenko, T, Barsukov, I. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras.
Nat. Cell Biol., 19, 2017
|
|
1ZVI
 
 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric-oxide synthase, brain, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
1L2E
 
 | | Human Kallikrein 6 (hK6) Active Form with benzamidine inhibitor | | Descriptor: | BENZAMIDINE, Kallikrein 6, MAGNESIUM ION | | Authors: | Bernett, M.J, Blaber, S.I, Scarisbrick, I.A, Blaber, M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and biochemical characterization of human kallikrein 6 reveals that a trypsin-like kallikrein is expressed in the central nervous system.
J.Biol.Chem., 277, 2002
|
|
8ALX
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, AMINOMETHYLAMIDE, ACETATE ION, ... | | Authors: | Rodriguez, I, Grudnik, P, Holak, T, Magiera-Mularz, K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biological characterization of pAC65, a macrocyclic peptide that blocks PD-L1 with equivalent potency to the FDA-approved antibodies.
Mol Cancer, 22, 2023
|
|
7UR2
 
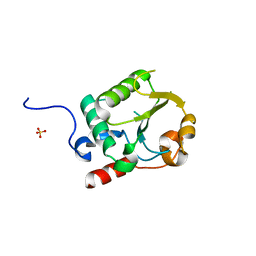 | |
4G79
 
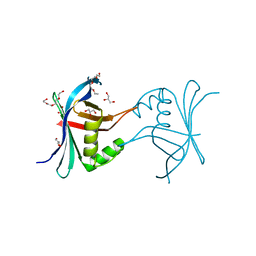 | | Structure a C. elegans SAS-6 variant | | Descriptor: | GLYCEROL, Spindle assembly abnormal protein 6, TETRAETHYLENE GLYCOL | | Authors: | Erat, M.C, Vakonakis, I. | | Deposit date: | 2012-07-20 | | Release date: | 2013-06-19 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1J08
 
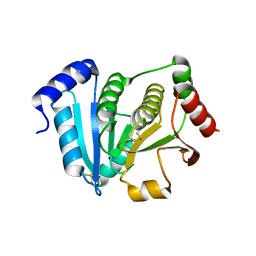 | | Crystal structure of glutaredoxin-like protein from Pyrococcus horikoshii | | Descriptor: | glutaredoxin-like protein | | Authors: | Tanaka, Y, Tanabe, E, Tsumoto, K, Kumagai, I, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-11 | | Release date: | 2003-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein disulfide isomerase from hyperthermophile as an additives of refolding of an immunoglobulin-folded protein
To be Published
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
8AVQ
 
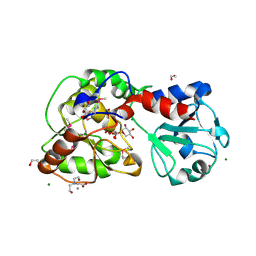 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AO75L in Complex with UDP-Xylose
To Be Published
|
|
1RJF
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, carboxy methyl transferase for protein phosphatase 2A catalytic subunit | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
4YHZ
 
 | | Crystal structure of 304M3-B Fab in complex with H3K4me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XBS
 
 | | Crystal structure of the adenylation domain of CmnG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CmnG | | Authors: | Chen, I.H, Wang, Y.L, Chang, C.Y. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.
Chembiochem, 23, 2022
|
|
7XBU
 
 | | Crystal structure of the adenylation domain of CmnG in complex with capreomycidine | | Descriptor: | (2S)-amino[(4R)-2-amino-1,4,5,6-tetrahydropyrimidin-4-yl]ethanoic acid, CmnG | | Authors: | Chen, I.H, Wang, Y.L, Chang, C.Y. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.
Chembiochem, 23, 2022
|
|
4DP6
 
 | | The 1.67 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 8.0 | | Descriptor: | COPPER (I) ION, GLYCEROL, Plastocyanin B, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4YHY
 
 | | Crystal structure of 309M3-B in complex with trimethylated Lys | | Descriptor: | Fab Heavy Chain, Fab Light Chain, N-TRIMETHYLLYSINE | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2PR1
 
 | | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A | | Descriptor: | COBALT (II) ION, COENZYME A, SULFATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Vorontsov, I.I, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-03 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Bacillus subtilis N-acetyltransferase YlbP protein in complex with Coenzyme-A.
To be Published
|
|
2PZ8
 
 | | NAD+ Synthetase from Bacillus anthracis with AMP-CPP and Mg2+ | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7UVA
 
 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7XBV
 
 | | Crystal structure of the adenylation domain of CmnG in complex with AMPCPP | | Descriptor: | CmnG, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PHOSPHATE ION | | Authors: | Chen, I.H, Wang, Y.L, Chang, C.Y. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.
Chembiochem, 23, 2022
|
|
