3N6U
 
 | |
1FKH
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
4BXK
 
 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with a domain-specific inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Douglas, R.G, Sharma, R.K, Masuyer, G, Lubbe, L, Zamora, I, Acharya, K.R, Chibale, K, Sturrock, E.D. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Design for the Development of N-Domain Selective Angiotensin-1 Converting Enzyme Inhibitors
Clin.Sci., 126, 2014
|
|
3UA1
 
 | |
2WJV
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, REGULATOR OF NONSENSE TRANSCRIPTS 2, SULFATE ION, ... | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
3TIK
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the tipifarnib derivative 6-((4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl)-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one | | Descriptor: | 6-[(R)-(4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl]-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha demethylase (CYP51) | | Authors: | Hargrove, T.Y, Wawrzak, Z, Kraus, J.M, Gelb, M.H, Buckner, F.S, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological characterization, structural studies, and in vivo activities of anti-chagas disease lead compounds derived from tipifarnib.
Antimicrob.Agents Chemother., 56, 2012
|
|
2WWD
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XAL
 
 | | Lead derivative of Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with ADP and IP6. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, ... | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5AVE
 
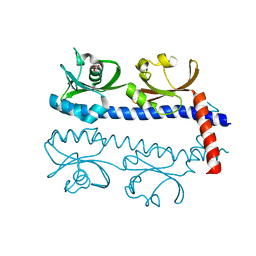 | | The ligand binding domain of Mlp37 with serine | | Descriptor: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
1DE6
 
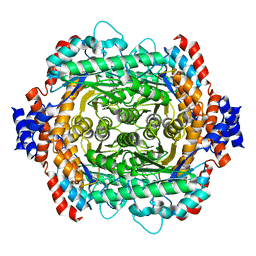 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNOSE, L-RHAMNOSE ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Korndorfer, I.P, Fessner, W.D, Matthews, B.W. | | Deposit date: | 1999-11-13 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1DFM
 
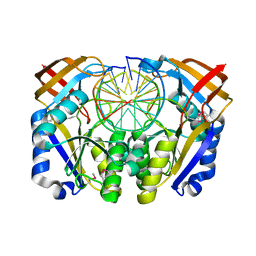 | | Crystal structure of restriction endonuclease BGLII complexed with DNA 16-mer | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-21 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
3U1N
 
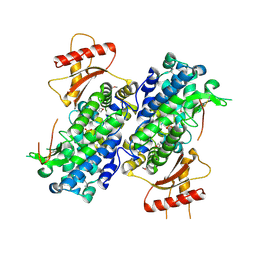 | | Structure of the catalytic core of human SAMHD1 | | Descriptor: | PHOSPHATE ION, SAM domain and HD domain-containing protein 1, ZINC ION | | Authors: | Goldstone, D.C, Ennis-Adeniran, V, Walker, P.A, Haire, L.F, Webb, M, Taylor, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase
Nature, 480, 2011
|
|
2WKK
 
 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | Descriptor: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | Deposit date: | 2009-06-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
1DLJ
 
 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
3NAH
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | RNA dependent RNA polymerase, SULFATE ION | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
2WIU
 
 | | Mercury-modified bacterial persistence regulator hipBA | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR HIPB, MERCURY (II) ION, ... | | Authors: | Evdokimov, A, Voznesensky, I, Fennell, K, Anderson, M, Smith, J.F, Fisher, D.A. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New Kinase Regulation Mechanism Found in Hipba: A Bacterial Persistence Switch.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3NC7
 
 | | CYP134A1 2-phenylimidazole bound structure | | Descriptor: | 2-phenyl-1H-imidazole, Cytochrome P450 cypX, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
3A60
 
 | | Crystal structure of unphosphorylated p70S6K1 (Form I) | | Descriptor: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
5A7O
 
 | | Crystal structure of human JMJD2A in complex with compound 42 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-(2-methoxyethanoylamino)-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5J4R
 
 | |
2Q8K
 
 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
3A8E
 
 | | The structure of AxCesD octamer complexed with cellopentaose | | Descriptor: | Cellulose synthase operon protein D, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Yao, M, Tanaka, I. | | Deposit date: | 2009-10-05 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1FOK
 
 | | STRUCTURE OF RESTRICTION ENDONUCLEASE FOKI BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*CP*TP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*T P*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*AP*G P*TP*CP*A)-3'), PROTEIN (FOKI RESTRICTION ENDONUCLEASE) | | Authors: | Aggarwal, D.A, Wah, J.A, Hirsch, L.F, Dorner, I, Schildkraut, A.K. | | Deposit date: | 1997-04-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the multimodular endonuclease FokI bound to DNA.
Nature, 388, 1997
|
|
5A2L
 
 | | Crystal structure of scFv-SM3 in complex with APD-CGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, MODIFIED ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3RSG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
