3OSF
 
 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-2f-13 DNA | | Descriptor: | 5'-D(*CP*AP*AP*GP*AP*CP*GP*AP*TP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*AP*TP*CP*GP*TP*CP*TP*TP*G)-3', ISOPROPYL ALCOHOL, ... | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
6WLU
 
 | | V. cholerae glycine riboswitch with glycine models, 5.7 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
5CEY
 
 | |
6WOS
 
 | |
3P3W
 
 | |
2BHZ
 
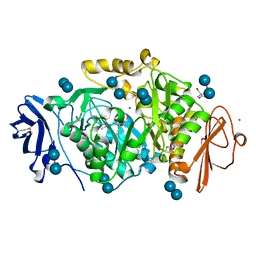 | | Crystal structure of Deinococcus radiodurans maltooligosyltrehalose trehalohydrolase in complex with maltose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, MAGNESIUM ION, ... | | Authors: | Timmins, J, Leiros, H.-K.S, Leonard, G, Leiros, I, McSweeney, S. | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Maltooligosyltrehalose Trehalohydrolase from Deinococcus Radiodurans in Complex with Disaccharides
J.Mol.Biol., 347, 2005
|
|
6PS4
 
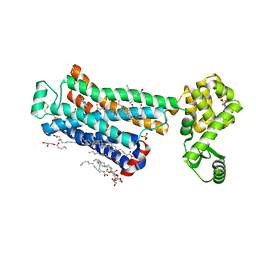 | | XFEL beta2 AR structure by ligand exchange from Timolol to ICI-118551. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
2BS8
 
 | | Crystal structure of F17b-G in complex with N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESIN | | Authors: | Buts, L, Wellens, A, VanMolle, I, Wyns, L, Loris, R, Lahmann, M, Oscarson, S, DeGreve, H, Bouckaert, J. | | Deposit date: | 2005-05-18 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of Natural Variation in Bacterial F17G Adhesins on Crystallization Behaviour.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5YER
 
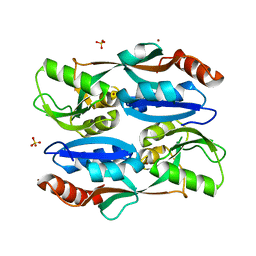 | | Regulatory domain of HypT from Salmonella typhimurium (Bromide ion-bound) | | Descriptor: | BROMIDE ION, Cell density-dependent motility repressor, SULFATE ION | | Authors: | Jo, I, Hong, S, Ahn, J, Ha, N.C. | | Deposit date: | 2017-09-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6WLK
 
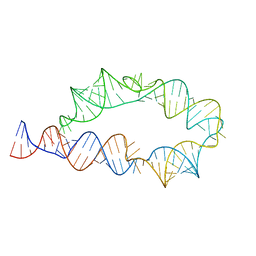 | | Apo ATP-TTR-3 models, 10.0 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6PY4
 
 | |
2XH1
 
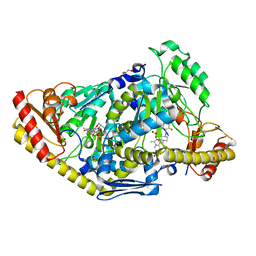 | | Crystal structure of human KAT II-inhibitor complex | | Descriptor: | (3S)-10-(4-AMINOPIPERAZIN-1-YL)-9-FLUORO-7-HYDROXY-3-METHYL-2,3-DIHYDRO-8H-[1,4]OXAZINO[2,3,4-IJ]QUINOLINE-6-CARBOXYLATE, GLYCEROL, IODIDE ION, ... | | Authors: | Rossi, F, Casazza, V, Garavaglia, S, Sathyasaikumar, K.V, Schwarcz, R, Kojima, S.I, Okuwaki, K, Ono, S.I, Kajii, Y, Rizzi, M. | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure-Based Selective Targeting of the Pyridoxal 5'-Phsosphate Dependent Enzyme Kynurenine Aminotransferase II for Cognitive Enhancement
J.Med.Chem., 53, 2010
|
|
5YIB
 
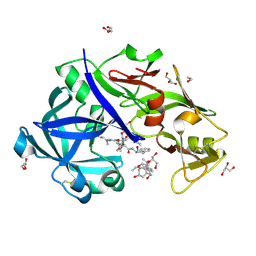 | | Crystal Structure of KNI-10743 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-(dimethylamino)ethyl-methyl-amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
6WUN
 
 | |
2C1A
 
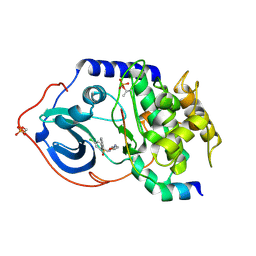 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
5CLL
 
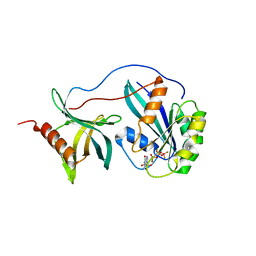 | | Truncated Ran wild type in complex with GDP-BeF and RanBD1 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
6TGF
 
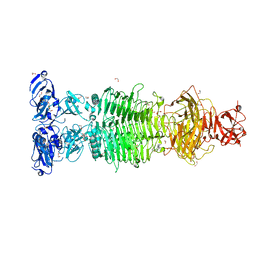 | | Pantoea stewartii WceF is a glycan biofilm modifying enzyme with a bacteriophage tailspike-like parallel beta-helix fold | | Descriptor: | 1,2-ETHANEDIOL, Exopolysaccharide biosynthesis protein, TETRAETHYLENE GLYCOL | | Authors: | Irmscher, T, Roske, Y, Gayk, I, Heinemann, U, Barbirz, S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold.
J.Biol.Chem., 296, 2021
|
|
2C5O
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
6TI4
 
 | | SHMT from Streptococcus thermophilus Tyr55Ser variant in complex with PLP/D-Serine/Lys230 gem diamine complex | | Descriptor: | (2~{R})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Y55S Serine Hydroxymethyltransferase variant from Streptococcus thermophilus in complex with gem-diamine intermediate of D-serine
To Be Published
|
|
2XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
6G5K
 
 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B in complex with human synaptotagmin 1 | | Descriptor: | Botulinum neurotoxin type B, Synaptotagmin-1 | | Authors: | Masuyer, G, Elliot, M, Favre-Guilmard, C, Liu, S.M, Maignel, J, Beard, M, Carre, D, Kalinichev, M, Lezmi, S, Mir, I, Nicoleau, C, Palan, S, Perier, C, Raban, E, Dong, M, Krupp, J, Stenmark, P. | | Deposit date: | 2018-03-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered botulinum neurotoxin B with improved binding to human receptors has enhanced efficacy in preclinical models.
Sci Adv, 5, 2019
|
|
6WLQ
 
 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
5C9F
 
 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | Descriptor: | Cake3, GLYCEROL, PHOSPHATE ION | | Authors: | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
