8RC4
 
 | | Structure of Integrator-PP2A complex | | Descriptor: | DSS1, Integrator complex subunit 1, Integrator complex subunit 10, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
4LDQ
 
 | |
4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
4LJ8
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2V5K
 
 | | Class II aldolase HpcH - magnesium - oxamate complex | | Descriptor: | 2,4-DIHYDROXYHEPT-2-ENE-1,7-DIOIC ACID ALDOLASE, MAGNESIUM ION, OXAMIC ACID, ... | | Authors: | Rea, D, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Mechanism of Hpch: A Metal Ion Dependent Class II Aldolase from the Homoprotocatechuate Degradation Pathway of Escherichia Coli.
J.Mol.Biol., 373, 2007
|
|
2V77
 
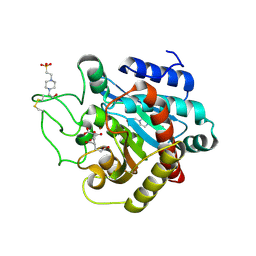 | | Crystal Structure of Human Carboxypeptidase A1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBOXYPEPTIDASE A1, OCTANE-1,3,5,7-TETRACARBOXYLIC ACID, ... | | Authors: | Pallares, I, Fernandez, D, Comellas-Bigler, M, Fernandez-Recio, J, Ventura, S, Aviles, F.X, Vendrell, J. | | Deposit date: | 2007-07-27 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Direct interaction between a human digestive protease and the mucoadhesive poly(acrylic acid).
Acta Crystallogr. D Biol. Crystallogr., D64, 2008
|
|
4L9J
 
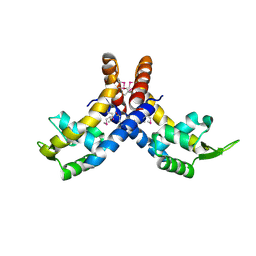 | |
4L9T
 
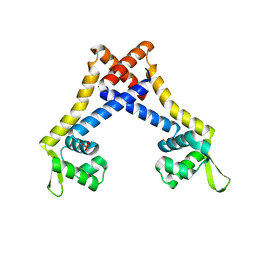 | |
7NE8
 
 | | Tick salivary protein BSAP1 | | Descriptor: | tick salivary protein BSAP1 | | Authors: | Denisov, S.S, Ippel, J.H, Castoldi, E, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of anticoagulant and anticomplement activity of the tick salivary protein Salp14 and its homologs.
J.Biol.Chem., 297, 2021
|
|
4LAS
 
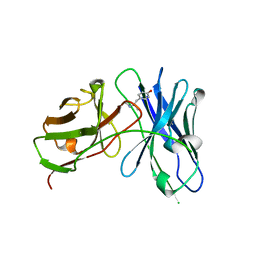 | | Crystal structure of a therapeutic single chain antibody in complex with 4-hydroxymethamphetamine | | Descriptor: | 4-[(2S)-2-(methylamino)propyl]phenol, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7N45
 
 | |
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
8RDJ
 
 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
4LNW
 
 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4LJ4
 
 | | ClpB NBD2 from T. thermophilus, nucleotide-free | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2V0K
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
7NL5
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1R,2R,3R,4S,5R)-4-(hydroxymethyl)cyclohexane-1,2,3,5-tetrol, (1R,6S)-5beta-(Hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2beta,3beta,4alpha-triol, Alpha-1,6-mannanase, ... | | Authors: | Schroeder, S, Offen, W.A, Males, A, Jin, Y, De Boer, C, Enotarpi, J, Marino, L, van der Marel, G.A, Florea, B.I, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
4LDL
 
 | | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-[(1R)-1-hydroxy-2-{[1-(4-hydroxyphenyl)-2-methylpropan-2-yl]amino}ethyl]benzene-1,2-diol, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
7RTM
 
 | | Cryo-EM Structure of the Sodium-driven Chloride/Bicarbonate Exchanger NDCBE (SLC4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Wang, W.G, Tsirulnikov, K, Zhekova, H, Kayik, G, Muhammad-Khan, H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the sodium-driven chloride/bicarbonate exchanger NDCBE.
Nat Commun, 12, 2021
|
|
4LJ9
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with AMPPCP | | Descriptor: | Chaperone protein ClpB, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LJN
 
 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
8RKQ
 
 | | Structure of human DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (ALDH4A1) complexed with the molecular tweezer CLR01 | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial | | Authors: | Porfetye, A.T, Vetter, I.R. | | Deposit date: | 2023-12-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Do Molecular Tweezers Bind to Proteins? Lessons from X-ray Crystallography.
Molecules, 29, 2024
|
|
8RKR
 
 | | Structure of human DELTA-1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE (ALDH4A1) complexed with a monophosphate-tweezer | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-22-(Dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaen-3-ol, BENZOIC ACID, Delta-1-pyrroline-5-carboxylate dehydrogenase, ... | | Authors: | Porfetye, A.T, Vetter, I.R. | | Deposit date: | 2023-12-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | How Do Molecular Tweezers Bind to Proteins? Lessons from X-ray Crystallography.
Molecules, 29, 2024
|
|
