1PXI
 
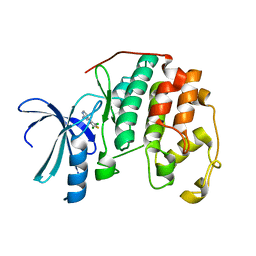 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,5-Dichloro-thiophen-3-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,5-DICHLOROTHIEN-3-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
3NIE
 
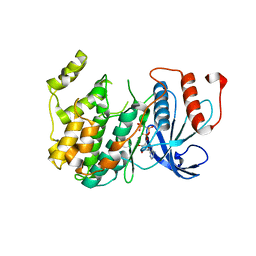 | | Crystal Structure of PF11_0147 | | Descriptor: | MAP2 kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Cossar, D, Amani, M, Artz, J.D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of PF11_0147
To be Published
|
|
3IU2
 
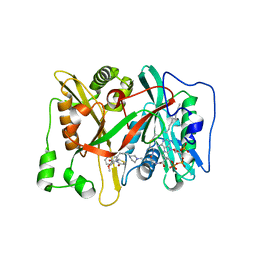 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096 | | Descriptor: | (2R)-2-{4-hydroxy-5-methoxy-2-[3-(4-methylpiperazin-1-yl)propyl]phenyl}-3-pyridin-3-yl-1,3-thiazolidin-4-one, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD90096
To be Published
|
|
1XW6
 
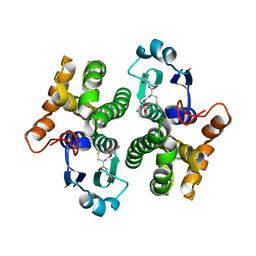 | | 1.9 angstrom resolution structure of human glutathione S-transferase M1A-1A complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-10-29 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
4KF3
 
 | | Crystal Structure of Myotoxin II (MjTX-II), a myotoxic Lys49-phospholipase A2 from Bothrops moojeni. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog 2, ISOPROPYL ALCOHOL | | Authors: | Salvador, G.H.M, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and functional studies with mytoxin II from Bothrops moojeni reveal remarkable similarities and differences compared to other catalytically inactive phospholipases A2-like.
Toxicon, 72C, 2013
|
|
1XZO
 
 | | Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins | | Descriptor: | CADMIUM ION, CALCIUM ION, Hypothetical protein ypmQ | | Authors: | Ye, Q, Imriskova-Sosova, I, Hill, B.C, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of a Disulfide Switch in BsSco, a Member of the Sco Family of Cytochrome c Oxidase Assembly Proteins
Biochemistry, 44, 2005
|
|
4KAT
 
 | | Crystal structure of FDTS from T. maritima mutant (R174K) with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Thymidylate synthase | | Authors: | Mathews, I.I. | | Deposit date: | 2013-04-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Flavin-Dependent Thymidylate Synthase as a Drug Target for Deadly Microbes: Mutational Study and a Strategy for Inhibitor Design.
J Bioterror Biodef, Suppl 12, 2013
|
|
5U0K
 
 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
1PW2
 
 | | APO STRUCTURE OF HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-06-30 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the
activation loop.
Structure, 11, 2003
|
|
5TPV
 
 | | X-ray structure of WlaRA (TDP-fucose-3,4-ketoisomerase) from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-DIPHOSPHATE, WlaRA, ... | | Authors: | Holden, H.M, Thoden, J.B, Li, Z.A, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinograd, E, Schoenhofen, I.C, Gilbert, M, Li, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5U3P
 
 | | Crystal Structure of DH511.4 Fab | | Descriptor: | DH511.4 Fab Heavy Chain, DH511.4 Fab Light Chain | | Authors: | Nicely, N.I, Williams, L.D, Ofek, G, Haynes, B.F. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and broad HIV-neutralizing antibodies in memory B cells and plasma.
Sci Immunol, 2, 2017
|
|
1P9A
 
 | |
5U9C
 
 | | 1.9 Angstrom Resolution Crystal Structure of dTDP-4-dehydrorhamnose Reductase from Yersinia enterocolitica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Olphie, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of dTDP-4-dehydrorhamnose Reductase from Yersinia enterocolitica.
To Be Published
|
|
4LZW
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymidine at 1.29 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 2016
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
1Z0E
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
4BKX
 
 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
5TUR
 
 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | 1-methyl-2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridine-4-carbonitrile, Serine/threonine-protein kinase pim-1 | | Authors: | Mechin, I, Zhang, Y, Wang, R, Batchelor, J.D, Mclean, L. | | Deposit date: | 2016-11-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1PXJ
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
2WNS
 
 | | Human Orotate phosphoribosyltransferase (OPRTase) domain of Uridine 5' -monophosphate synthase (UMPS) in complex with its substrate orotidine 5'-monophosphate (OMP) | | Descriptor: | CHLORIDE ION, OROTATE PHOSPHORIBOSYLTRANSFERASE, OROTIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotyenova, T, Kotzch, A, Nielsen, T.K, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-07-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Orotate Phosphoribosyltransferase (Oprtase) Domain of Uridine 5'-Monophosphate Synthase (Umps) in Complex with its Substrate Orotidine 5'-Monophosphate (Omp)
To be Published
|
|
1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
4B9K
 
 | | pVHL-ELOB-ELOC complex_(2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Buckley, D.L, Gustafson, J.L, VanMolle, I, Roth, A.G, SeopTae, H, Gareiss, P.C, Jorgensen, W.L, Ciulli, A, Crews, C.M. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1 Alpha
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4B20
 
 | | Structural basis of DNA loop recognition by Endonuclease V | | Descriptor: | 5'-D(*AP*TP*CP*TP*TP*GP*TP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*AP*CP*AP*GP)-3', ENDONUCLEASE V, ... | | Authors: | Rosnes, I, Rowe, A.D, Forstrom, R.J, Alseth, I, Bjoras, M, Dalhus, B. | | Deposit date: | 2012-07-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of DNA Loop Recognition by Endonuclease V.
Structure, 21, 2013
|
|
1Z0T
 
 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Dauter, Z, Botos, I, LaRonde-LeBlanc, N, Wlodawer, A. | | Deposit date: | 2005-03-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pathological crystallography: case studies of several unusual macromolecular crystals.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z9N
 
 | |
