8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
6HCU
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum bound to a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | Authors: | Tamjar, J, Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YT9
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) substrate-unbound. | | Descriptor: | GLYCEROL, Peptidylarginine deiminase, SODIUM ION | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
6MLK
 
 | | Structure of Thioesterase from DEBS with a thioesterase-specific antibody | | Descriptor: | 6-deoxyerythronolide-B synthase EryA3, modules 5 and 6, CHLORIDE ION, ... | | Authors: | Mathews, I.I, Li, X, Khosla, C. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery and Characterization of a Thioesterase-Specific Monoclonal Antibody That Recognizes the 6-Deoxyerythronolide B Synthase.
Biochemistry, 57, 2018
|
|
6PZF
 
 | |
8ESY
 
 | | D30N mutant HIV protease in complex with benzoxaborolone analog of darunavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Windsor, I.W, Graham, B.J, Raines, R.T. | | Deposit date: | 2022-10-15 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
5NJG
 
 | | Structure of an ABC transporter: part of the structure that could be built de novo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
3MF2
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bll0957 protein, GLYCEROL, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8F0F
 
 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
7AKJ
 
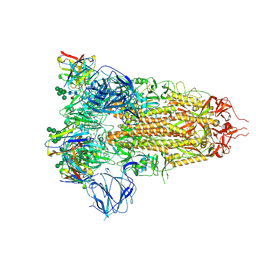 | | Structure of the SARS-CoV spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
4YNP
 
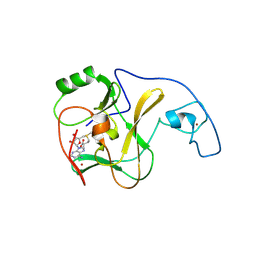 | | ASH1L SET domain S2259M mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.-J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-10 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
8EIP
 
 | |
7AIP
 
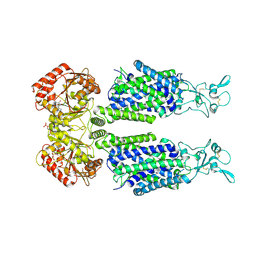 | | Structure of Human Potassium Chloride Transporter KCC1 in NaCl (Reference Map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ebenhoch, R, Chi, G, Man, H, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Liko, I, Tehan, B.G, Almeida, F.G, Elkins, J, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Tang, H, Robinson, C.V, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
8EIN
 
 | |
4GFC
 
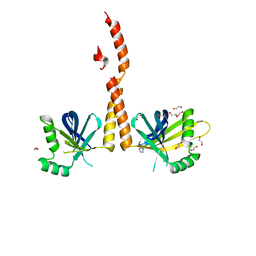 | | N-terminal coiled-coil dimer of C.elegans SAS-6, crystal form B | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Spindle assembly abnormal protein 6, ... | | Authors: | Erat, M.C, Vakonakis, I. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
8VMF
 
 | | Crystal structure of a transition-state mimic of the GSK-3/Axin complex bound to a beta-catenin S45D peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Axin-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
8VME
 
 | | Crystal structure of the GSK-3/Axin complex bound to a phosphorylated beta-catenin T41A peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Axin-1, Catenin beta-1, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
7ARW
 
 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
8VMG
 
 | | Crystal structure of GSK-3 26-383 bound to Axin 383-435 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
3MEY
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bll0957 protein, MAGNESIUM ION, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4GMS
 
 | | Crystal structure of heterosubtypic Fab S139/1 in complex with influenza A H3 hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S139/1 heavy chain, ... | | Authors: | Lee, P.S, Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7AF3
 
 | | Bacterial 30S ribosomal subunit assembly complex state M (head domain) | | Descriptor: | 16S rRNA (head), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S.R. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
