4YUI
 
 | | Multiconformer synchrotron model of CypA at 180 K | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | 登録日 | 2015-03-18 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4ENH
 
 | |
6JL3
 
 | |
6JLB
 
 | |
4YUK
 
 | | Multiconformer synchrotron model of CypA at 260 K | | 分子名称: | Peptidyl-prolyl cis-trans isomerase A | | 著者 | Keedy, D.A, Kenner, L.R, Warkentin, M, Woldeyes, R.A, Thompson, M.C, Brewster, A.S, Van Benschoten, A.H, Baxter, E.L, Hopkins, J.B, Uervirojnangkoorn, M, McPhillips, S.E, Song, J, Mori, R.A, Holton, J.M, Weis, W.I, Brunger, A.T, Soltis, M, Lemke, H, Gonzalez, A, Sauter, N.K, Cohen, A.E, van den Bedem, H, Thorne, R.E, Fraser, J.S. | | 登録日 | 2015-03-18 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography.
Elife, 4, 2015
|
|
4GXX
 
 | |
6N4O
 
 | |
6SMG
 
 | | Structure of Coxsackievirus A10 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2019-08-21 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6SVB
 
 | | Terahertz irradiated structure of bovine trypsin (odd frames of crystal x40) | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | 登録日 | 2019-09-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVJ
 
 | | Terahertz irradiated structure of bovine trypsin (odd frames of crystal x42) | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | 登録日 | 2019-09-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVR
 
 | | Reference structure of bovine trypsin (odd frames of crystal x28) | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | 登録日 | 2019-09-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
3CCN
 
 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | 分子名称: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | 著者 | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | 登録日 | 2008-02-26 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
4YJ6
 
 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | 分子名称: | Aryl acylamidase, PHOSPHATE ION | | 著者 | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | 登録日 | 2015-03-03 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
6JFQ
 
 | |
6SSX
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | 分子名称: | Alpha-synuclein | | 著者 | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | 登録日 | 2019-09-09 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
3C7L
 
 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | 分子名称: | Regulator of G-protein signaling 16 | | 著者 | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | 登録日 | 2008-02-07 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6N7M
 
 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | 分子名称: | Hypothetical Protein CD630_05490 | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-11-27 | | 公開日 | 2018-12-12 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
5KHI
 
 | | HCN2 CNBD in complex with purine riboside-3', 5'-cyclic monophosphate (cPuMP) | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Purine riboside-3',5'-cyclic monophosphate | | 著者 | Ng, L.C.T, Putrenko, I, Baronas, V, Van Petegem, F, Accili, E.A. | | 登録日 | 2016-06-14 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cyclic Purine and Pyrimidine Nucleotides Bind to the HCN2 Ion Channel and Variably Promote C-Terminal Domain Interactions and Opening.
Structure, 24, 2016
|
|
4WGH
 
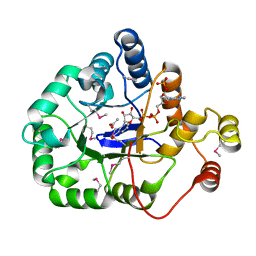 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | 分子名称: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-09-18 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
4WEN
 
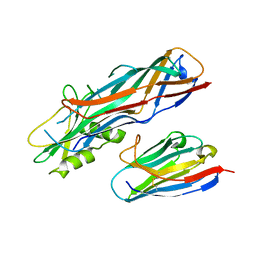 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V2 | | 分子名称: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC | | 著者 | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | 登録日 | 2014-09-10 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
6TJD
 
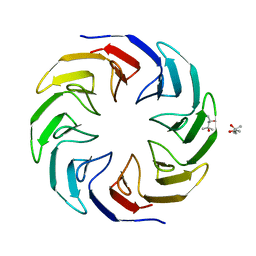 | | Crystal structure of the computationally designed Cake4 protein | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | 著者 | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
3BZ9
 
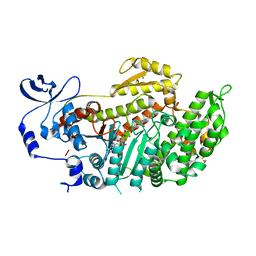 | | Crystal Structures of (S)-(-)-Blebbistatin Analogs bound to Dictyostelium discoideum myosin II | | 分子名称: | (3aS)-3a-hydroxy-1-phenyl-1,2,3,3a-tetrahydro-4H-pyrrolo[2,3-b]quinolin-4-one, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Allingham, J.S, Rayment, I. | | 登録日 | 2008-01-17 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The small molecule tool (S)-(-)-blebbistatin: novel insights of relevance to myosin inhibitor design.
Org.Biomol.Chem., 6, 2008
|
|
3BZZ
 
 | | Crystal structural of the mutated R313T EscU/SpaS C-terminal domain | | 分子名称: | EscU | | 著者 | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | 登録日 | 2008-01-18 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.407 Å) | | 主引用文献 | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
4IG7
 
 | |
4WI3
 
 | |
