2MTZ
 
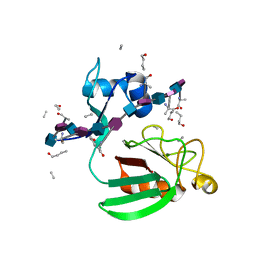 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
3QQB
 
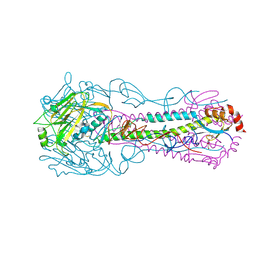 | | Crystal structure of HA2 R106H mutant of H2 hemagglutinin, neutral pH form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of an Early Fusion Intermediate of Influenza Virus Hemagglutinin.
J.Virol., 85, 2011
|
|
5F6F
 
 | |
1TF0
 
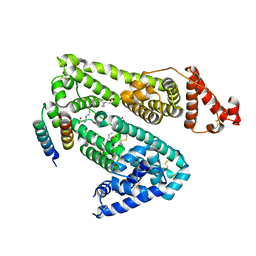 | | Crystal structure of the GA module complexed with human serum albumin | | Descriptor: | CITRIC ACID, DECANOIC ACID, Peptostreptococcal albumin-binding protein, ... | | Authors: | Lejon, S, Svensson, S, Bjorck, L, Frick, I.-M, Wikstrom, M. | | Deposit date: | 2004-05-26 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and biological implications of a bacterial albumin binding module in complex with human serum albumin
J.Biol.Chem., 279, 2004
|
|
1TFH
 
 | | EXTRACELLULAR DOMAIN OF HUMAN TISSUE FACTOR | | Descriptor: | HUMAN TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.G9 complex.
J.Mol.Biol., 275, 1998
|
|
3S2K
 
 | | Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahn, V.E, Chu, M.L.-H, Choi, H.-J, Tran, D, Abo, A, Weis, W.I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Wnt Signaling Inhibition by Dickkopf Binding to LRP5/6.
Dev.Cell, 21, 2011
|
|
1TKW
 
 | | The transient complex of poplar plastocyanin with turnip cytochrome f determined with paramagnetic NMR | | Descriptor: | COPPER (II) ION, Cytochrome f, HEME C, ... | | Authors: | Lange, C, Cornvik, T, Diaz-Moreno, I, Ubbink, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The transient complex of poplar plastocyanin with cytochrome f: effects of ionic strength and pH
Biochim.Biophys.Acta, 1707, 2005
|
|
3S3J
 
 | |
3VSE
 
 | | Crystal structure of methyltransferase | | Descriptor: | Putative uncharacterized protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kita, S, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2012-04-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of a putative methyltransferase SAV1081 from Staphylococcus aureus
Protein Pept.Lett., 20, 2012
|
|
6ERO
 
 | | Structure of human TFB2M | | Descriptor: | CHLORIDE ION, Dimethyladenosine transferase 2, mitochondrial,Dimethyladenosine transferase 2, ... | | Authors: | Hillen, H.S, Morozov, Y.I, Sarfallah, A, Temiakov, D, Cramer, P. | | Deposit date: | 2017-10-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Mitochondrial Transcription Initiation.
Cell, 171, 2017
|
|
1TF3
 
 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1TBA
 
 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | Descriptor: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | Authors: | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | Deposit date: | 1998-08-16 | | Release date: | 1999-08-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
3VH9
 
 | | Crystal structure of Aeromonas proteolytica aminopeptidase complexed with 8-quinolinol | | Descriptor: | Bacterial leucyl aminopeptidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Hanaya, K, Suetsugu, M, Kobayashi, K, Yamato, I, Aoki, S. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Potent inhibition of dinuclear zinc(II) peptidase, an aminopeptidase from Aeromonas proteolytica, by 8-quinolinol derivatives: inhibitor design based on Zn(2+) fluorophores, kinetic, and X-ray crystallographic study.
J.Biol.Inorg.Chem., 17, 2012
|
|
1TAM
 
 | | HUMAN IMMUNODEFICIENCY VIRUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 MATRIX PROTEIN | | Authors: | Matthews, S, Barlow, P, Clark, N, Kingsman, S, Kingsman, A, Campbell, I. | | Deposit date: | 1996-02-07 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of p17, the HIV matrix protein.
Biochem.Soc.Trans., 23, 1995
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
3VJM
 
 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
2MRF
 
 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2MOT
 
 | | Backbone Structure of Actin Depolymerizing Factor (ADF) of Toxoplasma gondii Based on Prot3DNMR Approach | | Descriptor: | Actin depolymerizing factor ADF | | Authors: | Kumar, D, Raikwal, N, Raval, I, Jaiswal, N, Shukla, V, Arora, A. | | Deposit date: | 2014-05-05 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Prot3DNMR: A Simple and Swift Strategy for Backbone Structure Determination of Proteins by NMR
To be Published
|
|
2MZA
 
 | |
2MPN
 
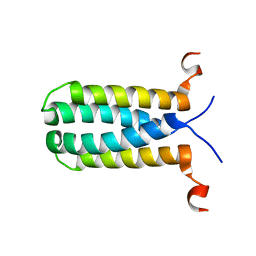 | | 3D NMR structure of the transmembrane domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
3RXA
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXK
 
 | | Crystal structure of Trypsin complexed with methyl 4-amino-1-methyl-pyrrolidine-2-carboxylate | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
1SWC
 
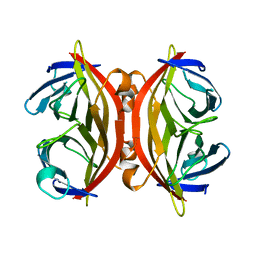 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1SWB
 
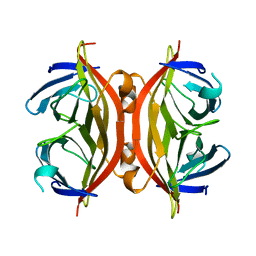 | | APO-CORE-STREPTAVIDIN AT PH 7.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
6EIO
 
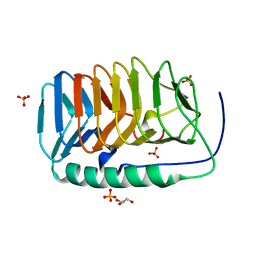 | | Crystal structure of an ice binding protein from an Antarctic Biological Consortium | | Descriptor: | Antifreeze protein, GLYCEROL, SULFATE ION | | Authors: | Nardini, M, Mangiagalli, M, Nardone, V, Bar Dolev, M, Vena, V.F, Sarusi, G, Braslavsky, I, Lotti, M. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Structure of a bacterial ice binding protein with two faces of interaction with ice.
FEBS J., 285, 2018
|
|
