6PVN
 
 | |
6PVL
 
 | |
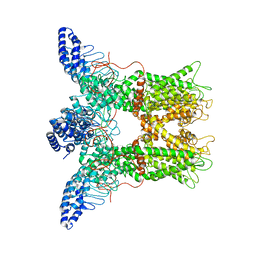
6PWS
 
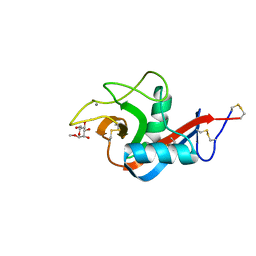 | |
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
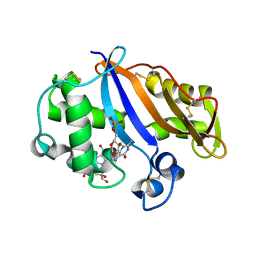
5NPJ
 
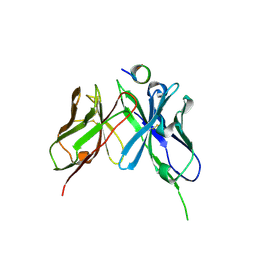 | |
6MDZ
 
 | |
4Z2S
 
 | | The crystal structure of Sclerotium Rolfsii lectin variant 2 (SSR2) in complex with N-acetyl-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
6SDX
 
 | | Salmonella ATPase InvC with ATP gamma S | | Descriptor: | ATP synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bernal, I, Roemermann, J, Flacht, L, Lunelli, M, Uetrecht, C, Kolbe, M. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural analysis of ligand-bound states of the Salmonella type III secretion system ATPase InvC.
Protein Sci., 28, 2019
|
|
6PVM
 
 | |
5NP2
 
 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NQX
 
 | | Structure of a fHbp(V1.1):PorA(P1.16) chimera. Fusion at fHbp position 294. | | Descriptor: | Factor H binding protein,Major outer membrane protein P.IA,Factor H binding protein | | Authors: | Johnson, S, Jongerius, I, Lea, S.M, Tang, C.M. | | Deposit date: | 2017-04-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structure-based design of chimeric antigens for multivalent protein vaccines.
Nat Commun, 9, 2018
|
|
1S5D
 
 | | Cholera holotoxin with an A-subunit Y30S mutation, Crystal form 2 | | Descriptor: | Cholera enterotoxin, A chain, GLYCEROL, ... | | Authors: | O'Neal, C.J, Amaya, E.I, Jobling, M.G, Holmes, R.K, Hol, W.G. | | Deposit date: | 2004-01-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of an intrinsically active cholera toxin
mutant yield insight into the toxin activation mechanism
Biochemistry, 43, 2004
|
|
4Z4F
 
 | | Human Argonaute2 Bound to t1-DAP Target RNA | | Descriptor: | MAGNESIUM ION, PHENOL, Protein argonaute-2, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
6QOY
 
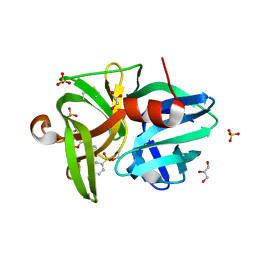 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-azanylethyl)benzenesulfonic acid, CHLORIDE ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kudryakova, I, Afoshin, A, Vasilyeva, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine bacteriolytic protease L1 of Lysobacter sp. XL1 complexed with protease inhibitor AEBSF: features of interaction
Process Biochem, 2019
|
|
4Z4D
 
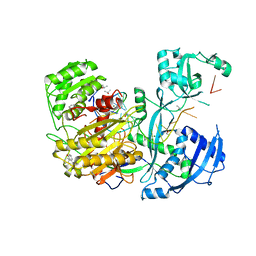 | | Human Argonaute2 Bound to t1-G Target RNA | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHENOL, ... | | Authors: | Schirle, N.T, MacRae, I.J. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.
Elife, 4, 2015
|
|
6PSE
 
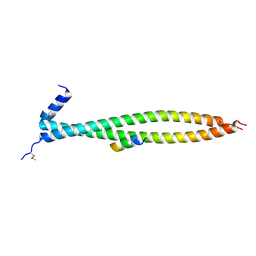 | |
6QNQ
 
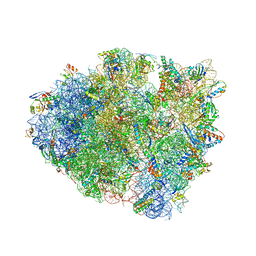 | | 70S ribosome initiation complex (IC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
4FWV
 
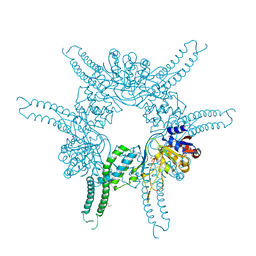 | | Crystal structure of the N-terminal domain of the Lon-like protease MtaLonC | | Descriptor: | SULFATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Li, J.K, Kuo, C.I, Huang, K.F. | | Deposit date: | 2012-07-02 | | Release date: | 2013-06-26 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The N-terminal substrate-recognition domain of a LonC protease exhibits structural and functional similarity to cytosolic chaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6FTB
 
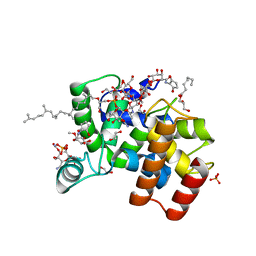 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | Authors: | Punekar, A.S, Dowson, C.J, Roper, D.I. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
1RXF
 
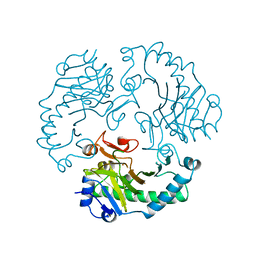 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
5N5C
 
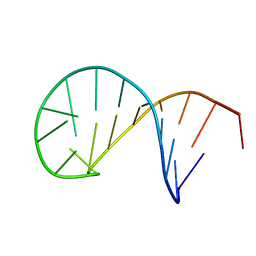 | | NMR solution structure of the TSL2 RNA hairpin | | Descriptor: | RNA (19-MER) | | Authors: | Garcia-Lopez, A, Wacker, A, Tessaro, F, Jonker, H.R.A, Richter, C, Comte, A, Berntenis, N, Schmucki, R, Hatje, K, Sciarra, D, Konieczny, P, Fournet, G, Faustino, I, Orozco, M, Artero, R, Goekjian, P, Metzger, F, Ebeling, M, Joseph, B, Schwalbe, H, Scapozza, L. | | Deposit date: | 2017-02-13 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes.
Nat Commun, 9, 2018
|
|
6PVU
 
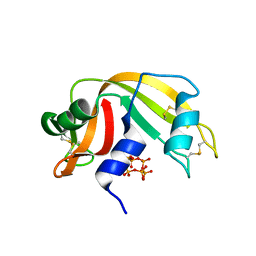 | | RNase A in complex with hexametaphosphate | | Descriptor: | 2,4,6,8,10,12-hexahydroxy-2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~,10lambda~5~,12lambda~5~-cyclohexaphosphoxane-2,4,6,8,10,12-hexone, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PVW
 
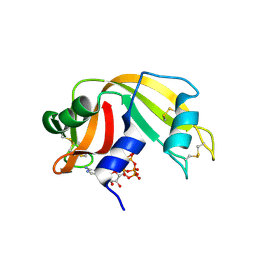 | | RNase A in complex with cp4pA | | Descriptor: | 2,4,6,8-tetrahydroxy-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocane-2,4,6,8-tetrone, 5'-O-[(R)-hydroxy{[(4R,8S)-4,6,8-trihydroxy-2,4,6,8-tetraoxo-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocan-2-yl]oxy}phosphoryl]adenosine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PWT
 
 | |
6ME1
 
 | |
