6WY4
 
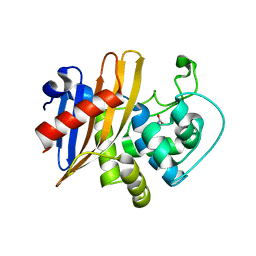 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
3UPC
 
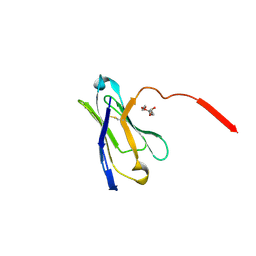 | | A general strategy for the generation of human antibody variable domains with increased aggregation resistance | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Dudgeon, K, Rouet, R, Kokmeijer, I, Langley, D.B, Christ, D. | | Deposit date: | 2011-11-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | General strategy for the generation of human antibody variable domains with increased aggregation resistance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LIO
 
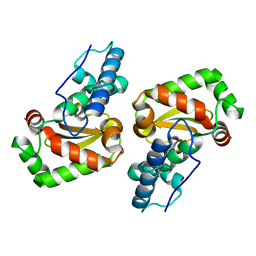 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
8HRH
 
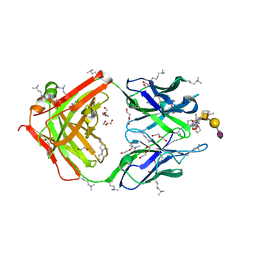 | | SN-131/1B2 anti-MUC1 antibody with a glycopeptide | | Descriptor: | 1-ACETYL-L-PROLINE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALANINE, ... | | Authors: | Wakui, H, Horidome, C, Yao, M, Ose, T, Nishimura, S.-I. | | Deposit date: | 2022-12-15 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and molecular insight into antibody recognition of dynamic neoepitopes in membrane tethered MUC1 of pancreatic cancer cells and secreted exosomes.
Rsc Chem Biol, 4, 2023
|
|
3CK8
 
 | | B. thetaiotaomicron SusD with beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
4OTX
 
 | | Structure of the anti-Francisella tularensis O-antigen antibody N203 Fab fragment | | Descriptor: | AZIDE ION, CHLORIDE ION, N203 heavy chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Roche, M.I, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Characterization of Francisella tularensis O-Antigen Antibodies at the Low End of Antigen Reactivity.
Monoclon Antib Immunodiagn Immunother, 33, 2014
|
|
3CK9
 
 | | B. thetaiotaomicron SusD with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SusD, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
7T2U
 
 | |
5KLN
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169A in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLK
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ and 2-hydroxymuconate-6-semialdehyde | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
4P2V
 
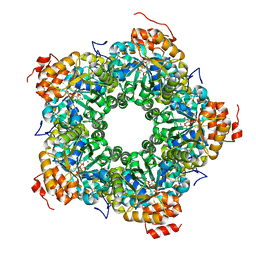 | | Structure of the AI-2 processing enzyme LsrF in complex with the product of the LsrG reaction P-HPD | | Descriptor: | (3R)-3-hydroxy-2,4-dioxopentyl dihydrogen phosphate, Uncharacterized aldolase LsrF | | Authors: | Miller, S.T, Oh, I.K, Xavier, K.B. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3LJF
 
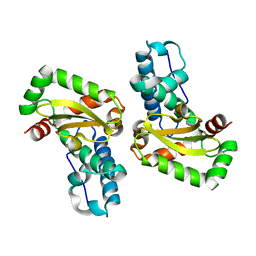 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
2C5X
 
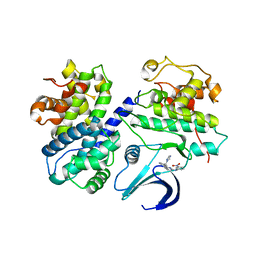 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, Mcinnes, C, Pandalaneni, S.R, Mcnae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
4V74
 
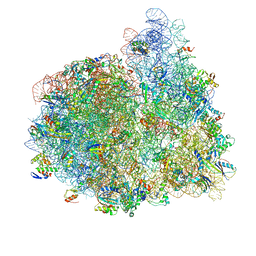 | | 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1S1Q
 
 | | TSG101(UEV) domain in complex with Ubiquitin | | Descriptor: | ACETIC ACID, COPPER (II) ION, SULFATE ION, ... | | Authors: | Sundquist, W.I, Schubert, H.L, Kelly, B.N, Hill, G.C, Holton, J.M, Hill, C.P. | | Deposit date: | 2004-01-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin recognition by the human TSG101 protein
Mol.Cell, 13, 2004
|
|
7SEH
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
5V7G
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
7SEI
 
 | | Glucose-6-phosphate 1-dehydrogenase (K403Q) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
1S59
 
 | | Structure of nucleoside diphosphate kinase 2 with bound dGTP from Arabidopsis | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Nucleoside diphosphate kinase II | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-01-20 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
4V7A
 
 | | E. coli 70S-fMetVal-tRNAVal post-translocation complex (post4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2BF4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
5VFB
 
 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
1S20
 
 | | A novel NAD binding protein revealed by the crystal structure of E. Coli 2,3-diketogulonate reductase (YiaK) NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82 | | Descriptor: | Hypothetical oxidoreductase yiaK, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
2C5V
 
 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, ALA-ALA-ABA-ARG-SER-LEU-ILE-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-02 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
4UXR
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
