3SXW
 
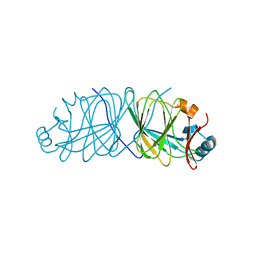 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
3THT
 
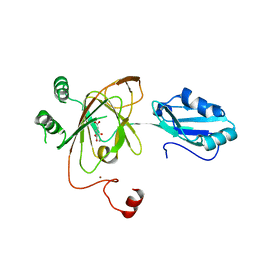 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3USH
 
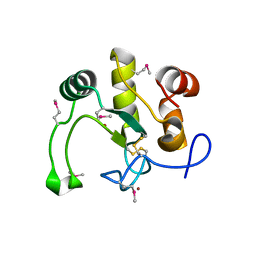 | | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207 | | Descriptor: | BROMIDE ION, Uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207
To be Published
|
|
1L2T
 
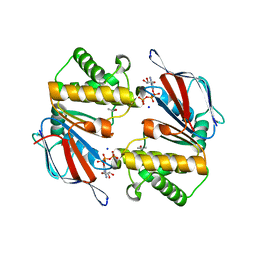 | | Dimeric Structure of MJ0796, a Bacterial ABC Transporter Cassette | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hypothetical ABC transporter ATP-binding protein MJ0796, ISOPROPYL ALCOHOL, ... | | Authors: | Smith, P.C, Karpowich, N, Rosen, J, Hunt, J.F. | | Deposit date: | 2002-02-24 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer.
Mol.Cell, 10, 2002
|
|
1L3I
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3B
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP W/ LONG CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1KXZ
 
 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
3SY1
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, UPF0001 protein yggS | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70
To be Published
|
|
3U19
 
 | | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | GLYCEROL, artificial protein OR51 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE
ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
3V45
 
 | | Crystal Structure of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium Target OR130 | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine hydrolase OSH55 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3W5W
 
 | | Mn2+-GMP complex of nanoRNase (Nrn) from Bacteroides fragilis | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Putative exopolyphosphatase-related protein | | Authors: | Uemura, Y, Nakagawa, N, Wakamatsu, T, Montelione, G.T, Hunt, J.F, Masui, R, Kuramitsu, S. | | Deposit date: | 2013-02-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins.
Febs Lett., 587, 2013
|
|
3U26
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR48 | | Descriptor: | PF00702 domain protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
1QYI
 
 | | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25. | | Descriptor: | hypothetical protein | | Authors: | Kuzin, A.P, Edstrom, W, Ma, L.C, Shin, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-10 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF Q8NW41 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ZR25.
To be Published
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UAK
 
 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
1PG6
 
 | | X-Ray Crystal Structure of Protein SPYM3_0169 from Streptococcus pyogenes. Northeast Structural Genomics Consortium Target DR2. | | Descriptor: | CALCIUM ION, Hypothetical protein SpyM3_0169 | | Authors: | Kuzin, A, Lee, I, Edstrom, W, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-05-27 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of hypothetical protein SPYM3_0169 from Streptococcus pyogenes
To be Published, 2003
|
|
3UW6
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR120 | | Descriptor: | Alanine racemase | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-08 | | Last modified: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
1SBK
 
 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
1SQ4
 
 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
1SPV
 
 | | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, putative polyprotein/phosphatase | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Phosphatase of Escherichia coli, Northeast Structural Genomoics Target ER58
To be Published
|
|
