1J8E
 
 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
2PGH
 
 | | STRUCTURE DETERMINATION OF AQUOMET PORCINE HEMOGLOBIN AT 2.8 ANGSTROM RESOLUTION | | Descriptor: | HEMOGLOBIN (AQUO MET) (ALPHA CHAIN), HEMOGLOBIN (AQUO MET) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Katz, D.S, White, S.P, Huang, W, Kumar, R, Christianson, D.W. | | Deposit date: | 1994-09-16 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of aquomet porcine hemoglobin at 2.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
3EZR
 
 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EZV
 
 | | CDK-2 with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
2B34
 
 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
3F5X
 
 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1ONR
 
 | | STRUCTURE OF TRANSALDOLASE B | | Descriptor: | TRANSALDOLASE B | | Authors: | Jia, J, Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-08-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of transaldolase B from Escherichia coli suggests a circular permutation of the alpha/beta barrel within the class I aldolase family.
Structure, 4, 1996
|
|
6UP7
 
 | | neurotensin receptor and arrestin2 complex | | Descriptor: | ARG-ARG-PRO-TYR-ILE-LEU, Beta-arrestin-1, Neurotensin receptor type 1, ... | | Authors: | Qu, Q.H, Huang, W, Masureel, M, Janetzko, J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-26 | | Last modified: | 2020-06-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the neurotensin receptor 1 in complex with beta-arrestin 1.
Nature, 579, 2020
|
|
8YRF
 
 | |
4IU9
 
 | | Crystal structure of a membrane transporter | | Descriptor: | Nitrite extrusion protein 2 | | Authors: | Yan, H, Huang, W, Yan, C, Gong, X, Jiang, S, Zhao, Y, Wang, J, Shi, Y. | | Deposit date: | 2013-01-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure and mechanism of a nitrate transporter.
Cell Rep, 3, 2013
|
|
4IU8
 
 | | Crystal structure of a membrane transporter (selenomethionine derivative) | | Descriptor: | NITRATE ION, Nitrite extrusion protein 2 | | Authors: | Yan, H, Huang, W, Yan, C, Gong, X, Jiang, S, Zhao, Y, Wang, J, Shi, Y. | | Deposit date: | 2013-01-20 | | Release date: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and mechanism of a nitrate transporter.
Cell Rep, 3, 2013
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3OXZ
 
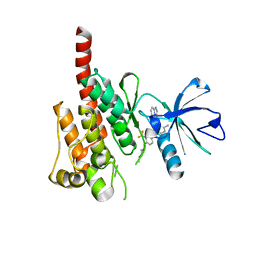 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
8FCK
 
 | | Structure of the vertebrate augmin complex | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | Authors: | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.88 Å) | | Cite: | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
8K6Z
 
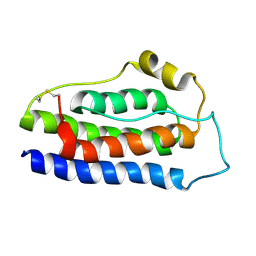 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
6JM5
 
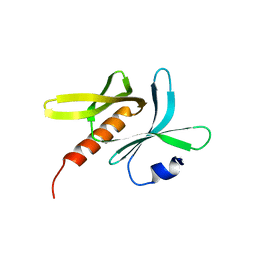 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2K3N
 
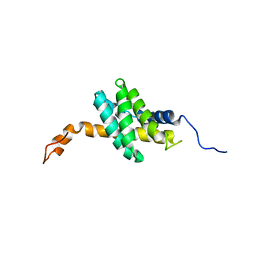 | |
2K3O
 
 | |
7T2H
 
 | | CryoEM structure of mu-opioid receptor - Gi protein complex bound to lofentanil (LFT) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Seven, A.B, Qu, Q, Huang, W, Robertson, M.J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2021-12-04 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into distinct signaling profiles of the mu OR activated by diverse agonists.
Nat.Chem.Biol., 2022
|
|
7TUY
 
 | | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | | Descriptor: | 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, G-protein coupled receptor 183,Soluble cytochrome b562 fusion, anti-BRIL Fab Heavy chain, ... | | Authors: | Chen, H, Huang, W, Li, X. | | Deposit date: | 2022-02-03 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
2K53
 
 | | NMR solution structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9 | | Descriptor: | A3DK08 Protein | | Authors: | Swapna, G.V.T, Huang, W, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of A3DK08 protein from Clostridium thermocellum: Northeast Structural Genomics Consortium Target CmR9
To be Published
|
|
6JJ7
 
 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
6JJ4
 
 | | Crystal structure of OsHXK6-apo form | | Descriptor: | Hexokinase-6 | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of OsHXK6-apo
To Be Published
|
|
