5G4G
 
 | | Structure of the ATPgS-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G4F
 
 | | Structure of the ADP-bound VAT complex | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Huang, R, Ripstein, Z.A, Augustyniak, R, Lazniewski, M, Ginalski, K, Kay, L.E, Rubinstein, J.L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Unfolding the Mechanism of the Aaa+ Unfoldase Vat by a Combined Cryo-Em, Solution NMR Study.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7MT0
 
 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTZ
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8W5J
 
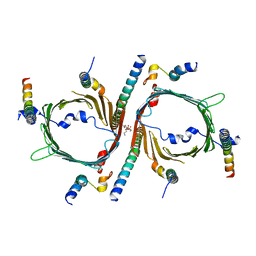 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
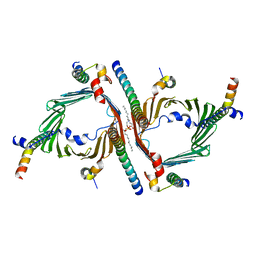 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
6UI7
 
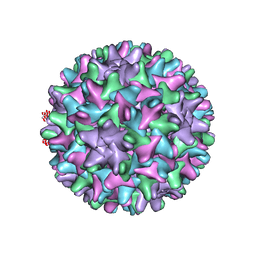 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6PVS
 
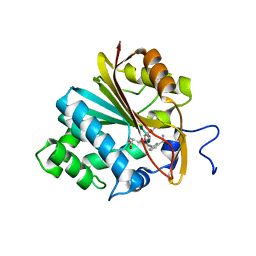 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL320 | | Descriptor: | 9-(5-{[(3R)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)prop-2-yn-1-yl]amino}-5-deoxy-alpha-D-lyxofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
6PVE
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor LL319 | | Descriptor: | 9-(5-{[(3S)-3-amino-3-carboxypropyl][3-(3-carbamoylphenyl)propyl]amino}-5-deoxy-alpha-D-ribofuranosyl)-9H-purin-6-amine, NNMT protein | | Authors: | Noinaj, N, Huang, R, Chen, D, Yadav, R. | | Deposit date: | 2019-07-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Propargyl-Linked Bisubstrate Analogues as Tight-Binding Inhibitors for NicotinamideN-Methyltransferase.
J.Med.Chem., 62, 2019
|
|
8TEX
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
8TEY
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
6PVB
 
 | | The structure of NTMT1 in complex with compound 6 | | Descriptor: | AMINO GROUP-()-(2~{S})-2-azanylpropanal-()-ISOLEUCINE-()-ARGININE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-9-(5-{[(3S)-3-amino-3-carboxypropyl](pentyl)amino}-5-deoxy-beta-L-arabinofuranosyl)-9H-purin-6-amine, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogues.
J.Med.Chem., 63, 2020
|
|
6PVA
 
 | | The structure of NTMT1 in complex with compound 11 | | Descriptor: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of NTMT1 in complex with compound 11
To Be published
|
|
8SHB
 
 | | Crystal Structure of PRMT3 with Compound YD1-208 | | Descriptor: | 5'-S-[3-(N'-phenylcarbamimidamido)propyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-208
To be published
|
|
8SIG
 
 | | Crystal Structure of PRMT4 with Compound YD1-288 | | Descriptor: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
8SIH
 
 | | Crystal Structure of PRMT4 with Compound YD1-289 | | Descriptor: | 5'-{[2-(benzylcarbamamido)ethyl][3-(N'-cyclopentylcarbamimidamido)propyl]amino}-5'-deoxyadenosine, CALCIUM ION, Histone-arginine methyltransferase CARM1 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-289
To be published
|
|
8SHR
 
 | | Crystal Structure of PRMT3 with Compound YD1-214 | | Descriptor: | 5'-S-[2-(phenylcarbamamido)ethyl]-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of PRMT3 with Compound YD1-214
To be published
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
8SII
 
 | | Crystal Structure of CBX7 with compound UNC4976 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Chromobox protein homolog 7, ... | | Authors: | Song, X, Dong, A, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of CBX7 with compound UNC4976
To be published
|
|
8DOK
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
7MTG
 
 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MUA
 
 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
