6YL8
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_011 | | Descriptor: | (2~{R},5~{S})-2-(2-chlorophenyl)-5-methyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNO
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6Y5P
 
 | | RING-DTC domain of Deltex1 bound to NAD | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase DTX1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
1O0L
 
 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
5TQX
 
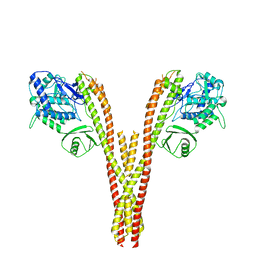 | | CryoEM reconstruction of human IKK1, intermediate conformation 2 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
6Y5N
 
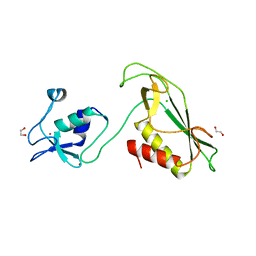 | | RING-DTC domain of Deltex1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase DTX1, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
5O6T
 
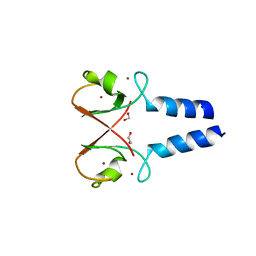 | | BIRC4 RING in complex with dimeric ubiquitin variant | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase XIAP, Polyubiquitin-B, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
5TQY
 
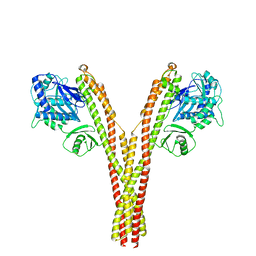 | | CryoEM reconstruction of human IKK1, closed conformation 3 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5O6S
 
 | |
5U01
 
 | | Cooperative DNA binding by two RelA dimers | | Descriptor: | DNA (27-MER), Transcription factor p65 | | Authors: | Ghosh, G, Huang, D. | | Deposit date: | 2016-11-22 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA-binding affinity and transcriptional activity of the RelA homodimer of nuclear factor kappa B are not correlated.
J. Biol. Chem., 292, 2017
|
|
5TQW
 
 | | CryoEM reconstruction of human IKK1, open conformation 1 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
4BHM
 
 | | The crystal structure of MoSub1-DNA complex reveals a novel DNA binding mode | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP)-3', MOSUB1 TRANSCRIPTION COFACTOR, SULFATE ION | | Authors: | Huang, J, Liu, H, Zhao, Y, Huang, D, liu, J, Peng, Y. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
1MY5
 
 | | NF-kappaB p65 subunit dimerization domain homodimer | | Descriptor: | NF-kappaB p65 (RelA) subunit | | Authors: | Huxford, T, Mishler, D, Phelps, C.B, Huang, D.-B, Sengchanthalangsy, L.L, Reeves, R, Hughes, C.A, Komives, E.A, Ghosh, G. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/I kappaB alpha complex formation
J.Mol.Biol., 324, 2002
|
|
1IKN
 
 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
1OY3
 
 | | CRYSTAL STRUCTURE OF AN IKBBETA/NF-KB P65 HOMODIMER COMPLEX | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Malek, S, Huang, D.B, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
5O76
 
 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
3BJL
 
 | |
1EK3
 
 | | KAPPA-4 IMMUNOGLOBULIN VL, REC | | Descriptor: | CALCIUM ION, CHLORIDE ION, KAPPA-4 IMMUNOGLOBULIN LIGHT CHAIN VL | | Authors: | Pokkuluri, P.R, Huang, D.-B, Raffen, R, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-03-06 | | Release date: | 2001-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of Amyloidogenic Kappa-4 Immunoglobulin VL, REC
To be Published
|
|
1K3Z
 
 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
2Y1M
 
 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6TTW
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 4 (ASI_M3M_047) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)-~{N}-piperidin-4-yl-oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TTT
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 2 (ASI_M3M_140) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-~{N}-methyl-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TTP
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 1/Adenosine (DHU_M3M_023) | | Descriptor: | ACETATE ION, ADENOSINE, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TTX
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 5 (ASI_M3M_051) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)-~{N}-(piperidin-4-ylmethyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TU1
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 8 (ASI_M3M_091) | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-2-[(6-aminopurin-9-yl)methyl]-5-azanyl-oxane-3,4-diol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-31 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
